Getting Started with R and RStudio
Jessica Minnier, PhD & Meike Niederhausen, PhD
OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop
2019/09/24 & 2020/02/19
slides: bit.ly/berd_intro_r
pdf: bit.ly/berd_intro_r_pdf
1. Open slides: bit.ly/berd_intro_r
2. Install R
- Windows:
- Download from https://cran.rstudio.com/bin/windows/base/
- Mac OS X:
- Download the latest .pkg file (currently R-3.6.2.pkg) from https://cran.rstudio.com/bin/macosx/
3. Install RStudio Desktop Open Source License
- Select download file corresponding to your operating system from https://www.rstudio.com/products/rstudio/download/#download
4. Download folder of data (unzip completely)
- Go to bit.ly/intro_rproj and unzip folder
- Open (double click on)
berd_intro_project.Rprojfile.
Questions
- Who has used R?
- What other statistical software have you used?
- Has anyone used other programming languages (C, java, python, etc)?
- Why do you want to learn R?
Learning Objectives
- Basic operations in R/RStudio
- Understand data structures
- Be able to load in data
- Basic operations on data
- Be able to make a plot
- Know how to get help
Introduction
Rrrrrr?
What is R?
- A programming language
- Focus on statistical modeling and data analysis
- import data, manipulate data, run statistics, make plots
- Useful for "Data Science"
- Great visualizations
- Also useful for most anything else you'd want to tell a computer to do
- Interfaces with other languages i.e. python, C++, bash

For the history and details: Wikipedia
- an interpreted language (run it through a command line)
- procedural programming with functions
- Why "R"?? Scheme (?) inspired S (invented at Bell Labs in 1976) which inspired R (free and open source! in 1992)
What is RStudio?
R is a programming language
RStudio is an integrated development environment (IDE) = an interface to use R (with perks!)
Modern Dive
Emma Rand
Rstudio demo
R Projects (why .Rproj file?) & Good Practices
Use projects to keep everything together (read this)
- Create an RStudio project for each data analysis project, for each homework assignment, etc.
- A project is associated with a directory folder
- Keep data files there
- Keep scripts there; edit them, run them in bits or as a whole
- Save your outputs (plots and cleaned data) there
- Only use relative paths, never absolute paths
- relative (good):
read.csv("data/mydata.csv") - absolute (bad):
read.csv("/home/yourname/Documents/stuff/mydata.csv")
- relative (good):
Advantages of using projects
- standardizes file paths
- keep everything together
- a whole folder can be easily shared and run on another computer
- when you open the project everything is as you left it
Let's code!
Coding in the console
Typing and execting code in the console
- Type code in the console
- Press return to execute the code
- Output shown below
Coding in the console is not advisable for most situations!
- We only recommend this for short pieces of code that you don't need to save
- We will be using scripts (
.Rfiles) to run and save code (in a few slides)
> 7[1] 7> 3 + 5[1] 8> "hello"[1] "hello"> # this is a comment, nothing happens> # 5 - 8> > # separate multiple commands with ;> 3 + 5; 4 + 8[1] 8[1] 12We can do math
> 10^2[1] 100> 3 ^ 7[1] 2187> 6/9[1] 0.6666667> 9-43[1] -34We can do math
> 10^2[1] 100> 3 ^ 7[1] 2187> 6/9[1] 0.6666667> 9-43[1] -34R follows the rules for order of operations and ignores spaces between numbers (or objects)
> 4^3-2* 7+9 /2[1] 54.5The equation above is computed as 43−(2⋅7)+92
Logarithms and exponentials
Logarithms: log() is base e
> log(10)[1] 2.302585> log10(10)[1] 1Logarithms and exponentials
Logarithms: log() is base e
> log(10)[1] 2.302585> log10(10)[1] 1Exponentials
> exp(1)[1] 2.718282> exp(0)[1] 1Logarithms and exponentials
Logarithms: log() is base e
> log(10)[1] 2.302585> log10(10)[1] 1Exponentials
> exp(1)[1] 2.718282> exp(0)[1] 1Check that log() is base e
> log(exp(1))[1] 1Using functions
log()is an example of a function- functions have "arguments"
?login console will show help forlog()
Arguments read in order:
> mean(1:4)[1] 2.5> seq(1,12,3)[1] 1 4 7 10Arguments read by name:
> mean(x = 1:4)[1] 2.5> seq(from = 1, to = 12, by = 3)[1] 1 4 7 10Variables
Data, information, everything is stored as a variable
- Can assign a variable using either
=or<-- Using
<-is preferable - type name of variable to print
- Using
Assigning just one value:
> x = 5> x[1] 5> x <- 5> x[1] 5Variables
Data, information, everything is stored as a variable
- Can assign a variable using either
=or<-- Using
<-is preferable - type name of variable to print
- Using
Assigning just one value:
> x = 5> x[1] 5> x <- 5> x[1] 5Assigning a vector of values
- Consecutive integers
> a <- 3:10> a[1] 3 4 5 6 7 8 9 10- Concatenate a string of numbers
> b <- c(5, 12, 2, 100, 8)> b[1] 5 12 2 100 8We can do math with variables
Math using variables with just one value
> x <- 5> x[1] 5> x + 3[1] 8> y <- x^2> y[1] 25We can do math with variables
Math using variables with just one value
> x <- 5> x[1] 5> x + 3[1] 8> y <- x^2> y[1] 25Math on vectors of values: element-wise computation
> a <- 3:6> a[1] 3 4 5 6> a+2; a*3[1] 5 6 7 8[1] 9 12 15 18> a*a[1] 9 16 25 36Variable can include text (characters)
> hi <- "hello"> hi[1] "hello"> greetings <- c("Guten Tag", "Hola", hi)> greetings[1] "Guten Tag" "Hola" "hello"Missing values
Missing values are denoted as NA and are handled differently depending on the operation. There are special functions for NA (i.e. is.na(), na.omit()).
> x <- c(1, 2, NA, 5)> is.na(x)[1] FALSE FALSE TRUE FALSE> mean(x)[1] NA> mean(x, na.rm=TRUE)[1] 2.666667> x <- c("a", "a", NA, "b")> table(x)xa b 2 1> table(x, useNA = "always")x a b <NA> 2 1 1Viewing list of defined variables
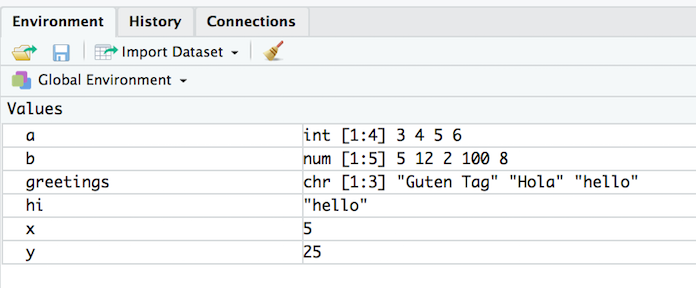
ls()is the R command to see what objects have been defined.- This list includes all defined objects (including dataframes, functions, etc.)
> ls()[1] "a" "b" "greetings" "hi" "x" "y"- You can also look at the list in the Environment window:
Removing defined variables
- The R command to delete an object is
rm().
> ls()[1] "a" "b" "greetings" "hi" "x" "y"> rm("greetings", hi) # Can run with or without quotes> ls()[1] "a" "b" "x" "y"- Remove EVERYTHING - Be careful!!
> rm(list=ls())> ls()character(0)- Can also remove everything using the Clear Workspace option in the Session menu.
Common console errors (1/2)
Incomplete commands
- When the console is waiting for a new command, the prompt line begins with
>- If the console prompt is
+, then a previous command is incomplete - You can finish typing the command in the console window
- If the console prompt is
Example:
> 3 + (2*6+ )[1] 15Common console errors (2/2)
Object is not found
- This happens when text is entered for a non-existent variable (object)
Example:
> helloError in eval(expr, envir, enclos): object 'hello' not found- Can be due to missing quotes
> install.packages(dplyr) # need install.packages("dplyr")Error in install.packages(dplyr): object 'dplyr' not foundR scripts (save your work!)
Coding in a script (1/3)
- Create a new script by
- selecting
File -> New File -> R Script, - or clicking on
 (the left most button at the top of the scripting window), and then selecting the first option
(the left most button at the top of the scripting window), and then selecting the first option R Script
- selecting
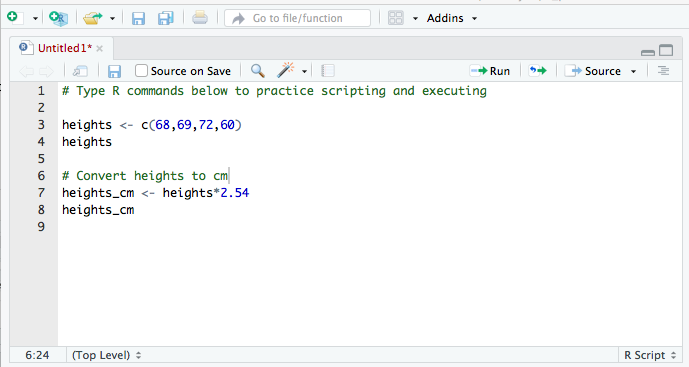
- Type code in the script
- Type each R command on its own line
- Use
#to convert text to comments so that text doesn't accidentally get executed as an R command
Coding in a script (2/3)
- Select code you want to execute, by
- placing the cursor in the line of code you want to execute,
- or highlighting the code you want to execute
- Execute code in the script, by
- clicking on the
 button in the top right corner of the scripting window,
button in the top right corner of the scripting window, - or typing one of the following key combinations to execute the code
- Windows: ctrl + return
- Mac: command + return
- clicking on the
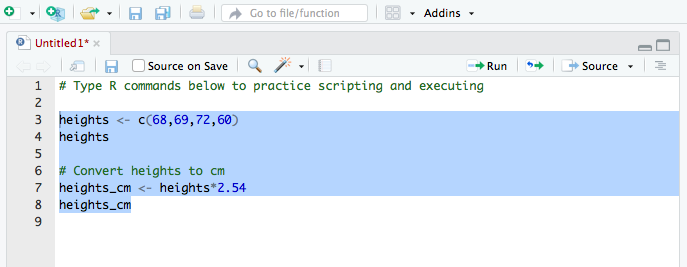
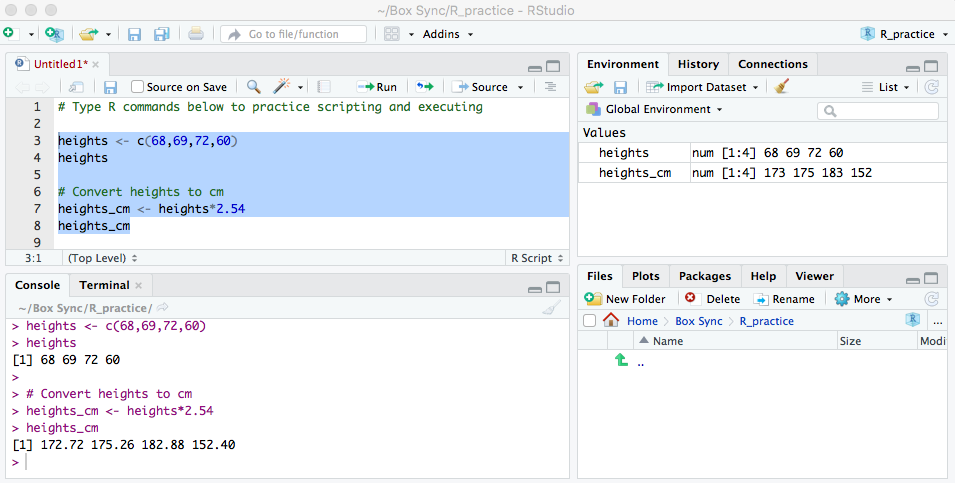
Coding in a script (3/3)
- The screenshot below shows code in the scripting window (top left window)
- The executed highlighted code and its output appear in the console window (bottom left window)
Useful keyboard shortcuts
| action | mac | windows/linux |
|---|---|---|
| run code in script | cmd + enter | ctrl + enter |
<- |
option + - | alt + - |
Try typing (with shortcut) in a script and running
y <- 5yNow, in the console, press the up arrow.
Others: (see full list)
| action | mac | windows/linux |
|---|---|---|
| interrupt currently executing command | esc | esc |
| in console, go to previously run code | up/down | up/down |
| keyboard shortcut help | option + shift + k | alt + shift + k |
Saving a script
Save a script by
- selecting
File -> Save, - or clicking on
 (towards the left above the scripting window)
(towards the left above the scripting window)
- selecting
You will need to specify
- a filename to save the script as
- ALWAYS use .R as the filename extension for R scripts
- the folder to save the script in
- a filename to save the script as
Practice time!
Practice 1
Open a new R script and type code/answers for next tasks in it. Save as
Practice1.RCreate a vector of all integers from 4 to 10, and save it as
a1.Create a vector of even integers from 4 to 10, and save it as
a2.What is the sum of
a1anda2?What does the command
sum(a1)do?What does the command
length(a1)do?Use the
sumandlengthcommands to calculate the average of the values ina1.Compute the sum of all integers from 1 to 100. Then compare your answer to the one you get using the formula for sum of the first n integers: n(n+1)/2.
Compute the sum of the squares of all integers from 1 to 100.
Take a break!
Object types
Data frames
Vectors vs. data frames: a data frame is a collection (or array or table) of vectors
df <- data.frame( IDs=1:3, gender=c("male", "female", "Male"), age=c(28, 35.5, 31), trt = c("control", "1", "1"), Veteran = c(FALSE, TRUE, TRUE) )df## IDs gender age trt Veteran## 1 1 male 28.0 control FALSE## 2 2 female 35.5 1 TRUE## 3 3 Male 31.0 1 TRUE- Allows different columns to be of different data types (i.e. numeric vs. text)
Both numeric and text can be stored within a column (stored together as text).
Vectors and data frames are examples of objects in R.
- There are other types of R objects to store data, such as matrices, lists, and tibbles.
- These will be discussed in future R workshops.
Variable (column) types
| type | description |
|---|---|
| integer | integer-valued numbers |
| numeric | numbers that are decimals |
| factor | categorical variables stored with levels (groups) |
| character | text, "strings" |
| logical | boolean (TRUE, FALSE) |
- View the structure of our data frame to see what the variable types are:
str(df)## 'data.frame': 3 obs. of 5 variables:## $ IDs : int 1 2 3## $ gender : Factor w/ 3 levels "female","male",..: 2 1 3## $ age : num 28 35.5 31## $ trt : Factor w/ 2 levels "1","control": 2 1 1## $ Veteran: logi FALSE TRUE TRUEData frame cells, rows, or columns
Show whole data frame
df## IDs gender age trt Veteran## 1 1 male 28.0 control FALSE## 2 2 female 35.5 1 TRUE## 3 3 Male 31.0 1 TRUESpecific cell value:
DatSetName[row#, column#]
# Second row, Third columndf[2, 3]## [1] 35.5Entire column:
DatSetName[, column#]
# Third columndf[, 3]## [1] 28.0 35.5 31.0Entire row: DatSetName[row#, ]
# Second rowdf[2,]## IDs gender age trt Veteran## 2 2 female 35.5 1 TRUEGetting the data into Rstudio
Load a data set
- Read in csv file from file path with code (filepath relative to Rproj directory)
mydata <- read.csv("data/yrbss_demo.csv")- Or, open saved file using Import Dataset button in Environment window:
 .
. - If you use this option, then copy and paste the importing code to your script so that you have a record of from where and how you loaded the data set.
View(mydata) # Can also view the data by clicking on its name in the Environment tab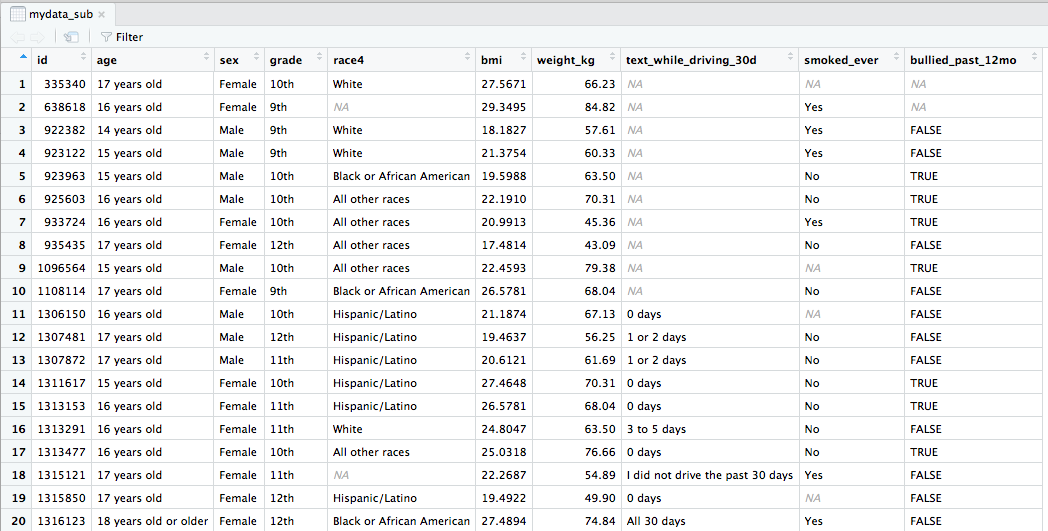
About the data
Data from the CDC's Youth Risk Behavior Surveillance System (YRBSS)
- small subset (20 rows) of the full complex survey data
- national school-based survey conducted by CDC and state, territorial, tribal, and local surveys conducted by state, territorial, and local education and health agencies and tribal governments
- monitors health-related behaviors (including alcohol & drug use, unhealthy & dangerous behaviors, sexuality, physical activity); see Questionnaires
- original data in the R package
yrbsswhich includes YRBSS from 1991-2013

Data set summary
summary(mydata)## id age sex grade ## Min. : 335340 14 years old :1 Female:12 10th:8 ## 1st Qu.: 925193 15 years old :4 Male : 8 11th:4 ## Median :1207132 16 years old :7 12th:4 ## Mean :1093150 17 years old :7 9th :4 ## 3rd Qu.:1313188 18 years old or older:1 ## Max. :1316123 ## race4 bmi weight_kg ## All other races :5 Min. :17.48 Min. :43.09 ## Black or African American:3 1st Qu.:20.36 1st Qu.:57.27 ## Hispanic/Latino :6 Median :22.23 Median :64.86 ## White :4 Mean :23.01 Mean :64.09 ## NA's :2 3rd Qu.:26.58 3rd Qu.:70.31 ## Max. :29.35 Max. :84.82 ## text_while_driving_30d smoked_ever bullied_past_12mo## 0 days : 5 No :10 Mode :logical ## 1 or 2 days : 2 Yes : 6 FALSE:11 ## 3 to 5 days : 1 NA's: 4 TRUE :7 ## All 30 days : 1 NA's :2 ## I did not drive the past 30 days: 1 ## NA's :10Data set info
dim(mydata)## [1] 20 10nrow(mydata)## [1] 20ncol(mydata)## [1] 10names(mydata)## [1] "id" "age" "sex" ## [4] "grade" "race4" "bmi" ## [7] "weight_kg" "text_while_driving_30d" "smoked_ever" ## [10] "bullied_past_12mo"Data structure
- What are the different variable types in this data set?
str(mydata) # structure of data## 'data.frame': 20 obs. of 10 variables:## $ id : int 335340 638618 922382 923122 923963 925603 933724 935435 1096564 1108114 ...## $ age : Factor w/ 5 levels "14 years old",..: 4 3 1 2 2 3 3 4 2 4 ...## $ sex : Factor w/ 2 levels "Female","Male": 1 1 2 2 2 2 1 1 2 1 ...## $ grade : Factor w/ 4 levels "10th","11th",..: 1 4 4 4 1 1 1 3 1 4 ...## $ race4 : Factor w/ 4 levels "All other races",..: 4 NA 4 4 2 1 1 1 1 2 ...## $ bmi : num 27.6 29.3 18.2 21.4 19.6 ...## $ weight_kg : num 66.2 84.8 57.6 60.3 63.5 ...## $ text_while_driving_30d: Factor w/ 5 levels "0 days","1 or 2 days",..: NA NA NA NA NA NA NA NA NA NA ...## $ smoked_ever : Factor w/ 2 levels "No","Yes": NA 2 2 2 1 1 2 1 NA 1 ...## $ bullied_past_12mo : logi NA NA FALSE FALSE TRUE TRUE ...View the beginning of a data set
head(mydata)## id age sex grade race4 bmi weight_kg## 1 335340 17 years old Female 10th White 27.5671 66.23## 2 638618 16 years old Female 9th <NA> 29.3495 84.82## 3 922382 14 years old Male 9th White 18.1827 57.61## 4 923122 15 years old Male 9th White 21.3754 60.33## 5 923963 15 years old Male 10th Black or African American 19.5988 63.50## 6 925603 16 years old Male 10th All other races 22.1910 70.31## text_while_driving_30d smoked_ever bullied_past_12mo## 1 <NA> <NA> NA## 2 <NA> Yes NA## 3 <NA> Yes FALSE## 4 <NA> Yes FALSE## 5 <NA> No TRUE## 6 <NA> No TRUEView the end of a data set
tail(mydata)## id age sex grade race4 bmi## 15 1313153 16 years old Female 11th Hispanic/Latino 26.5781## 16 1313291 16 years old Female 11th White 24.8047## 17 1313477 16 years old Female 10th All other races 25.0318## 18 1315121 17 years old Female 11th <NA> 22.2687## 19 1315850 17 years old Female 12th Hispanic/Latino 19.4922## 20 1316123 18 years old or older Female 12th Black or African American 27.4894## weight_kg text_while_driving_30d smoked_ever bullied_past_12mo## 15 68.04 0 days No TRUE## 16 63.50 3 to 5 days No FALSE## 17 76.66 0 days No TRUE## 18 54.89 I did not drive the past 30 days Yes FALSE## 19 49.90 0 days <NA> FALSE## 20 74.84 All 30 days Yes FALSESpecify how many rows to view at beginning or end of a data set
head(mydata, 3)## id age sex grade race4 bmi weight_kg## 1 335340 17 years old Female 10th White 27.5671 66.23## 2 638618 16 years old Female 9th <NA> 29.3495 84.82## 3 922382 14 years old Male 9th White 18.1827 57.61## text_while_driving_30d smoked_ever bullied_past_12mo## 1 <NA> <NA> NA## 2 <NA> Yes NA## 3 <NA> Yes FALSEtail(mydata, 1)## id age sex grade race4 bmi## 20 1316123 18 years old or older Female 12th Black or African American 27.4894## weight_kg text_while_driving_30d smoked_ever bullied_past_12mo## 20 74.84 All 30 days Yes FALSEWorking with the data
The $
Suppose we want to single out the column of BMI values.
- How did we previously learn to do this?
The $
Suppose we want to single out the column of BMI values.
- How did we previously learn to do this?
mydata[, 6]## [1] 27.5671 29.3495 18.1827 21.3754 19.5988 22.1910 20.9913 17.4814 22.4593## [10] 26.5781 21.1874 19.4637 20.6121 27.4648 26.5781 24.8047 25.0318 22.2687## [19] 19.4922 27.4894The problem with this method, is that we need to know the column number which can change as we make changes to the data set.
The $
Suppose we want to single out the column of BMI values.
- How did we previously learn to do this?
mydata[, 6]## [1] 27.5671 29.3495 18.1827 21.3754 19.5988 22.1910 20.9913 17.4814 22.4593## [10] 26.5781 21.1874 19.4637 20.6121 27.4648 26.5781 24.8047 25.0318 22.2687## [19] 19.4922 27.4894The problem with this method, is that we need to know the column number which can change as we make changes to the data set.
- Use the
$instead:DatSetName$VariableName
mydata$bmi## [1] 27.5671 29.3495 18.1827 21.3754 19.5988 22.1910 20.9913 17.4814 22.4593## [10] 26.5781 21.1874 19.4637 20.6121 27.4648 26.5781 24.8047 25.0318 22.2687## [19] 19.4922 27.4894Basic plots of numeric data: Histogram
hist(mydata$bmi)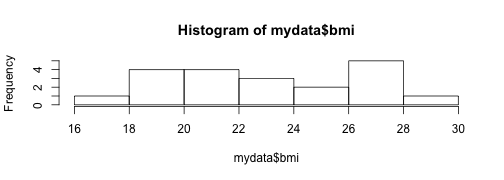
With extra features:
hist(mydata$bmi, xlab = "BMI", main="BMIs of students")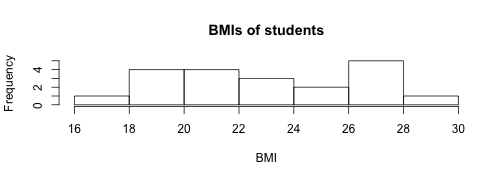
Basic plots of numeric data: Boxplot
boxplot(mydata$bmi)
Basic plots of numeric data: Boxplot
boxplot(mydata$bmi)
boxplot(mydata$bmi ~ mydata$sex, horizontal = TRUE, xlab = "BMI", ylab = "sex", main = "BMIs of students by sex")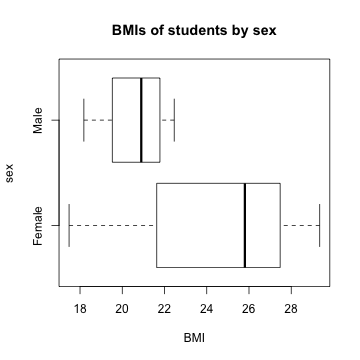
Basic plots of numeric data: Scatterplot
plot(mydata$weight_kg, mydata$bmi)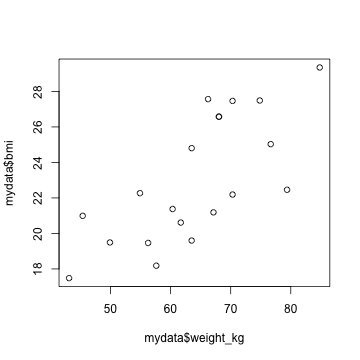
plot(mydata$weight_kg, mydata$bmi, xlab = "weight (kg)", ylab = "BMI", main = "BMI vs. Weight")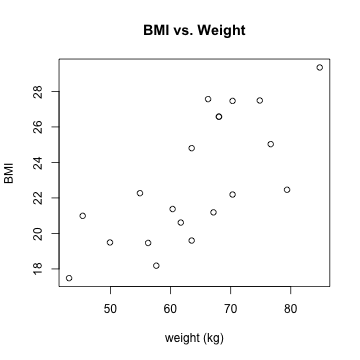
Summary stats of numeric data (1/2)
- Standard R
summarycommand
summary(mydata$bmi)## Min. 1st Qu. Median Mean 3rd Qu. Max. ## 17.48 20.36 22.23 23.01 26.58 29.35- Mean and standard deviation
mean(mydata$bmi)## [1] 23.00838sd(mydata$bmi)## [1] 3.56471Summary stats of numeric data (2/2)
- Min, max, & median
min(mydata$bmi)## [1] 17.4814max(mydata$bmi)## [1] 29.3495median(mydata$bmi)## [1] 22.22985- Quantiles
quantile(mydata$bmi, prob=c(0, .25, .5, .75, 1))## 0% 25% 50% 75% 100% ## 17.48140 20.35878 22.22985 26.57810 29.34950Add height column to data frame
Since BMI=kgm2, we have height(m)=√weight(kg)BMI
mydata$height_m <- sqrt( mydata$weight_kg / mydata$bmi )mydata$height_m## [1] 1.550000 1.699999 1.779999 1.680001 1.799998 1.780000 1.469998 1.570002## [9] 1.879998 1.600001 1.779998 1.699999 1.730001 1.600001 1.600001 1.600000## [17] 1.750001 1.569998 1.599999 1.650001dim(mydata)## [1] 20 11names(mydata)## [1] "id" "age" "sex" ## [4] "grade" "race4" "bmi" ## [7] "weight_kg" "text_while_driving_30d" "smoked_ever" ## [10] "bullied_past_12mo" "height_m"Access specific columns in data set
Previously we used DatSetName[, column#]
mydata[, c(2, 6)] # 2nd & 6th columns## age bmi## 1 17 years old 27.5671## 2 16 years old 29.3495## 3 14 years old 18.1827## 4 15 years old 21.3754## 5 15 years old 19.5988## 6 16 years old 22.1910## 7 16 years old 20.9913## 8 17 years old 17.4814## 9 15 years old 22.4593## 10 17 years old 26.5781## 11 16 years old 21.1874## 12 17 years old 19.4637## 13 17 years old 20.6121## 14 15 years old 27.4648## 15 16 years old 26.5781## 16 16 years old 24.8047## 17 16 years old 25.0318## 18 17 years old 22.2687## 19 17 years old 19.4922## 20 18 years old or older 27.4894The code below uses column names instead of numbers.
mydata[, c("age", "bmi")]## age bmi## 1 17 years old 27.5671## 2 16 years old 29.3495## 3 14 years old 18.1827## 4 15 years old 21.3754## 5 15 years old 19.5988## 6 16 years old 22.1910## 7 16 years old 20.9913## 8 17 years old 17.4814## 9 15 years old 22.4593## 10 17 years old 26.5781## 11 16 years old 21.1874## 12 17 years old 19.4637## 13 17 years old 20.6121## 14 15 years old 27.4648## 15 16 years old 26.5781## 16 16 years old 24.8047## 17 16 years old 25.0318## 18 17 years old 22.2687## 19 17 years old 19.4922## 20 18 years old or older 27.4894Access specific rows in data set
- Rows for 14 year olds only
mydata[mydata$age == "14 years old",] # 1 row since there is only one 14 year old## id age sex grade race4 bmi weight_kg text_while_driving_30d## 3 922382 14 years old Male 9th White 18.1827 57.61 <NA>## smoked_ever bullied_past_12mo height_m## 3 Yes FALSE 1.779999- Rows for teens with BMI less than 19
mydata[mydata$bmi < 19,]## id age sex grade race4 bmi weight_kg## 3 922382 14 years old Male 9th White 18.1827 57.61## 8 935435 17 years old Female 12th All other races 17.4814 43.09## text_while_driving_30d smoked_ever bullied_past_12mo height_m## 3 <NA> Yes FALSE 1.779999## 8 <NA> No FALSE 1.570002Access specific values in data set
- Grade and race for 15 year olds only
mydata[mydata$age == "15 years old", c("age", "grade", "race4")]## age grade race4## 4 15 years old 9th White## 5 15 years old 10th Black or African American## 9 15 years old 10th All other races## 14 15 years old 10th Hispanic/Latino- Age, sex, and BMI for students with BMI less than 19
mydata[mydata$bmi < 19, c("age", "sex", "bmi")]## age sex bmi## 3 14 years old Male 18.1827## 8 17 years old Female 17.4814Practice 2
Create a new script and save it as
Practice2.RCreate data frames for males and females separately.
Do males and females have similar BMIs? Weights? Compares means, standard deviations, range, and boxplots.
Plot BMI vs. weight for each gender separately. Do they have similar relationships?
Are males or females more likely to be bullied in the past 12 months? Calculate the percentage bullied for each gender.
Save data frame
- Save .RData file: the standard R format, which is recommended if saving data for future use in R
save(mydata, file = "data/mydata.RData") # saving mydata within the data folderYou can load .RData files using the load() command:
load("data/mydata.RData")- Save csv file: comma-separated values
write.csv(mydata, file = "data/mydata.csv", col.names = TRUE, row.names = FALSE)The more you know
Installing and using packages
- Packages are to R like apps are to your phone/OS
- Packages contain additional functions and data
- Install packages with
install.packages()- Also can use the "Packages" tab in Files/Plots/Packages/Help/Viewer window
- Only install once (unless you want to update)
- Installs from Comprehensive R Archive Network (CRAN) = package mothership
install.packages("dplyr") # only do this ONCE, use quotes- Load packages: At the top of your script include
library()commands to load each required package every time you open Rstudio.
library(dplyr) # run this every time you open Rstudio- Use a function without loading the package with
::
dplyr::arrange(mydata, bmi)## id age sex grade race4 bmi## 1 935435 17 years old Female 12th All other races 17.4814## 2 922382 14 years old Male 9th White 18.1827## 3 1307481 17 years old Male 12th Hispanic/Latino 19.4637## 4 1315850 17 years old Female 12th Hispanic/Latino 19.4922## 5 923963 15 years old Male 10th Black or African American 19.5988## 6 1307872 17 years old Male 11th Hispanic/Latino 20.6121## 7 933724 16 years old Female 10th All other races 20.9913## 8 1306150 16 years old Male 10th Hispanic/Latino 21.1874## 9 923122 15 years old Male 9th White 21.3754## 10 925603 16 years old Male 10th All other races 22.1910## 11 1315121 17 years old Female 11th <NA> 22.2687## 12 1096564 15 years old Male 10th All other races 22.4593## 13 1313291 16 years old Female 11th White 24.8047## 14 1313477 16 years old Female 10th All other races 25.0318## 15 1108114 17 years old Female 9th Black or African American 26.5781## 16 1313153 16 years old Female 11th Hispanic/Latino 26.5781## 17 1311617 15 years old Female 10th Hispanic/Latino 27.4648## 18 1316123 18 years old or older Female 12th Black or African American 27.4894## 19 335340 17 years old Female 10th White 27.5671## 20 638618 16 years old Female 9th <NA> 29.3495## weight_kg text_while_driving_30d smoked_ever bullied_past_12mo## 1 43.09 <NA> No FALSE## 2 57.61 <NA> Yes FALSE## 3 56.25 1 or 2 days No FALSE## 4 49.90 0 days <NA> FALSE## 5 63.50 <NA> No TRUE## 6 61.69 1 or 2 days No FALSE## 7 45.36 <NA> Yes TRUE## 8 67.13 0 days <NA> FALSE## 9 60.33 <NA> Yes FALSE## 10 70.31 <NA> No TRUE## 11 54.89 I did not drive the past 30 days Yes FALSE## 12 79.38 <NA> <NA> TRUE## 13 63.50 3 to 5 days No FALSE## 14 76.66 0 days No TRUE## 15 68.04 <NA> No FALSE## 16 68.04 0 days No TRUE## 17 70.31 0 days No TRUE## 18 74.84 All 30 days Yes FALSE## 19 66.23 <NA> <NA> NA## 20 84.82 <NA> Yes NA## height_m## 1 1.570002## 2 1.779999## 3 1.699999## 4 1.599999## 5 1.799998## 6 1.730001## 7 1.469998## 8 1.779998## 9 1.680001## 10 1.780000## 11 1.569998## 12 1.879998## 13 1.600000## 14 1.750001## 15 1.600001## 16 1.600001## 17 1.600001## 18 1.650001## 19 1.550000## 20 1.699999Installing packages from other places (i.e. github, URLs)
- Need to have remotes package installed first:
install.packages("remotes")- To install a package from github (often in development) use
install_github()from the remotes package
# https://github.com/hadley/yrbssremotes::install_github("hadley/yrbss")# Load it the same waylibrary(yrbss)How to get help (1/2)
Use ? in front of function name in console. Try this:
How to get help (2/2)
- Use
??(i.e??dplyror??read_csv) for searching all documentation in installed packages (including unloaded packages) - search Stack Overflow #r tag
- googlequestion + rcran or + r (i.e. "make a boxplot rcran" "make a boxplot r")
- google error in quotes (i.e.
"Evaluation error: invalid type (closure) for variable '***'") - search github for your function name (to see examples) or error
- Rstudio community
- twitter #rstats
Resources
- Click on this List of resources for learning R
- Watch recordings of our other workshops
- Highly recommend Data Wrangling in R with Tidyverse
Getting started:
- RStudio IDE Cheatsheet
- Install R/RStudio help video
- Basic Basics
Some of this is drawn from materials in online books/lessons:
- Intro to R/RStudio by Emma Rand
- Modern Dive - An Introduction to Statistical and Data Sciences via R by Chester Ismay & Albert Kim
- Cookbook for R by Winston Chang
Local resources
- OHSU's BioData club + active slack channel
- Portland's R user meetup group + active slack channel
- R-ladies PDX meetup group
- Cascadia R Conf - May 31, 2020 in Eugene with workshops
Contact info:
Jessica Minnier: minnier@ohsu.edu
Meike Niederhausen: niederha@ohsu.edu
This workshop info:
- Code for these slides on github: jminnier/berd_r_courses
- all the R code in an R script
- answers to practice problems can be found here: html, pdf
- The project folder of examples can be downloaded at github.com/jminnier/berd_intro_project & the solutions are in the
solns/folder.