An Introduction to R and RStudio for Exploratory Data Analysis
Jessica Minnier, PhD & Meike Niederhausen, PhD
OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop
Part 1: 2020/09/16 & Part 2: 2020/09/17
slides: bit.ly/berd_intro_part2
pdf: bit.ly/berd_intro_part2_pdf
An Introduction to R and RStudio for Exploratory Data Analysis (Part 2)
Instructors: Meike Niederhausen, PhD & Jessica Minnier, PhD
OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop
Do this now:
- Open html slides: http://bit.ly/berd_intro_part2
- Download Rmd file to follow along: bit.ly/berd_intro_rmd
- Open google doc for asking questions: bit.ly/berd_doc2
- Helpers will be monitoring this, you can ask questions, copy code or screenshots.
- Open Rstudio with these steps:
- Open the folder from yesterday
- Double click on the
.Rprojfile. - All your files should be there.
Working with data, continued
- Open your old Rmd file
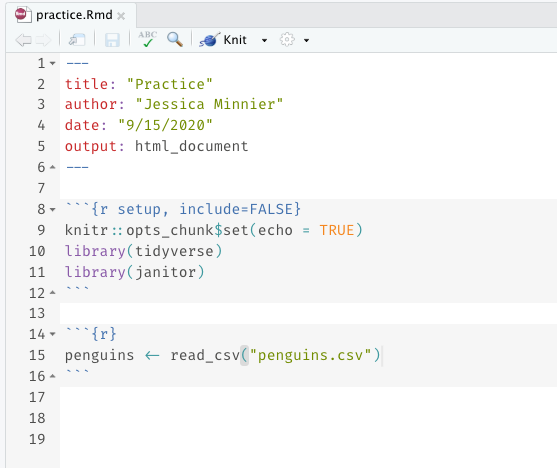
- If you make a new Rmd file, make sure you have this code in a code chunk at the top of the Rmd:
library(tidyverse)library(janitor)penguins <- read_csv("penguins")- Remember we need to load (open) the package every time we want to use it in a new Rstudio instance or knit an Rmd
- When you knit an Rmd, it is blind to what you have done in the console or in the R enivronment. It starts completely from scratch.
Working with data, we will use the pipe %>%
The pipe operator %>% is part of the tidyverse, and strings together commands to be performed sequentially
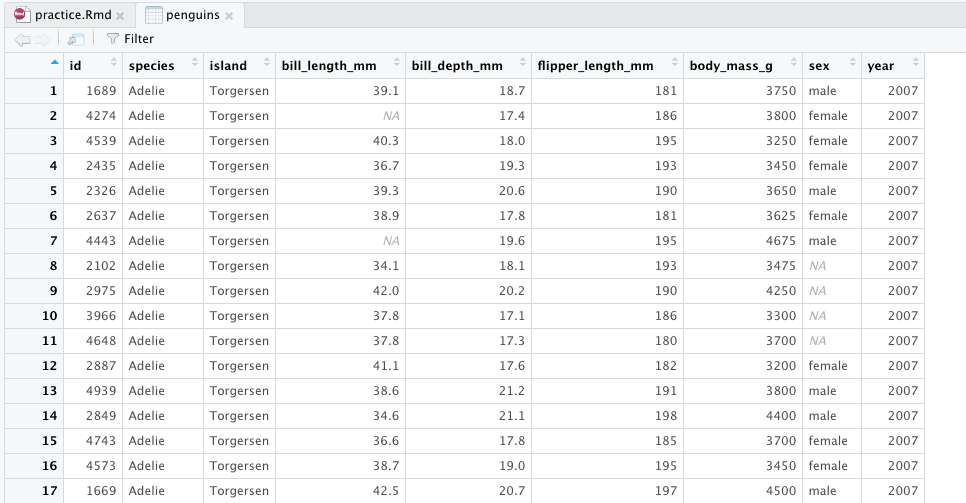
penguins %>% head(n=3) # prounounce %>% as "then"## # A tibble: 3 x 9## id species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g## <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl>## 1 1689 Adelie Torge… 39.1 18.7 181 3750## 2 4274 Adelie Torge… NA 17.4 186 3800## 3 4539 Adelie Torge… 40.3 18 195 3250## # … with 2 more variables: sex <chr>, year <dbl>- Always first list the tibble that the commands are being applied to
- Can use multiple pipes to run multiple commands in sequence
- What does the following code do?
penguins %>% head(n=2) %>% summary()Numerical data summaries: $ vs summarize()
We saw how to summarize a vector pulled with $, but there are easier ways to summarize multiple columns at once.
mean(penguins$body_mass_g)## [1] 4201.754median(penguins$body_mass_g)## [1] 4050penguins %>% summarize(mean(body_mass_g), median(body_mass_g))## # A tibble: 1 x 2## `mean(body_mass_g)` `median(body_mass_g)`## <dbl> <dbl>## 1 4202. 4050summarize() with NA
- Don't forget
na.rm = TRUEif you need it. - You can also name these columns.
penguins %>% summarize(mean_mass = mean(body_mass_g), mean_len = mean(bill_length_mm, na.rm = TRUE))## # A tibble: 1 x 2## mean_mass mean_len## <dbl> <dbl>## 1 4202. 44.0By group summarize() (1/2)
- We can summarize data as a whole, or in groups with
group_by() group_by()is very powerful, see data wrangling cheatsheet
# summary of all data as a wholepenguins %>% summarize(mass_mean =mean(body_mass_g), mass_sd = sd(body_mass_g), mass_cv = sd(body_mass_g)/mean(body_mass_g))## # A tibble: 1 x 3## mass_mean mass_sd mass_cv## <dbl> <dbl> <dbl>## 1 4202. 802. 0.191By group summarize() (2/2)
- We can summarize data as a whole, or in groups with
group_by() group_by()is very powerful, see data wrangling cheatsheet
# summary by group variablepenguins %>% group_by(species) %>% summarize(n_per_group = n(), mass_mean =mean(body_mass_g), mass_sd = sd(body_mass_g), mass_cv = sd(body_mass_g)/mean(body_mass_g))## # A tibble: 3 x 5## species n_per_group mass_mean mass_sd mass_cv## <chr> <int> <dbl> <dbl> <dbl>## 1 Adelie 151 3701. 459. 0.124 ## 2 Chinstrap 68 3733. 384. 0.103 ## 3 Gentoo 123 5076. 504. 0.0993Advanced summarize(across()) (1/3)
- Can also use
across()to summarize multiple variables (more examples)
penguins %>% summarize(across(c(body_mass_g, bill_depth_mm), mean))## # A tibble: 1 x 2## body_mass_g bill_depth_mm## <dbl> <dbl>## 1 4202. 17.2penguins %>% summarize(across(c(bill_length_mm, bill_depth_mm), mean, na.rm=TRUE))## # A tibble: 1 x 2## bill_length_mm bill_depth_mm## <dbl> <dbl>## 1 44.0 17.2Advanced summarize(across()) (2/3)
- Can also use
across()to summarize multiple variables and functions (more examples)
penguins %>% summarize(across(c(body_mass_g, bill_depth_mm), c(m = mean, sd = sd)))## # A tibble: 1 x 4## body_mass_g_m body_mass_g_sd bill_depth_mm_m bill_depth_mm_sd## <dbl> <dbl> <dbl> <dbl>## 1 4202. 802. 17.2 1.97Advanced summarize(across()) (3/3)
- Can also use
across()to summarize based on true/false conditions (more examples)

Allison Horst
penguins %>% summarize( across(where(is.character), n_distinct))## # A tibble: 1 x 3## species island sex## <int> <int> <int>## 1 3 3 3penguins %>% summarize(across(where(is.numeric), min, na.rm=TRUE))## # A tibble: 1 x 6## id bill_length_mm bill_depth_mm flipper_length_mm body_mass_g year## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 1001 32.1 13.1 172 2700 2007Frequency tables: simple count()
penguins %>% count(island)## # A tibble: 3 x 2## island n## <chr> <int>## 1 Biscoe 167## 2 Dream 124## 3 Torgersen 51penguins %>% count(species, island)## # A tibble: 5 x 3## species island n## <chr> <chr> <int>## 1 Adelie Biscoe 44## 2 Adelie Dream 56## 3 Adelie Torgersen 51## 4 Chinstrap Dream 68## 5 Gentoo Biscoe 123Fancier frequency tables: janitor package's tabyl function
# default tablepenguins %>% tabyl(species)## species n percent## Adelie 151 0.4415205## Chinstrap 68 0.1988304## Gentoo 123 0.3596491# output can be treated as tibblepenguins%>%tabyl(species)%>%select(-n)## species percent## Adelie 0.4415205## Chinstrap 0.1988304## Gentoo 0.3596491adorn_ your table!
penguins %>% tabyl(species) %>% adorn_totals("row") %>% adorn_pct_formatting(digits=2)## species n percent## Adelie 151 44.15%## Chinstrap 68 19.88%## Gentoo 123 35.96%## Total 342 100.00%2x2 tabyls
# default 2x2 tablepenguins %>% tabyl(species, sex)## species female male NA_## Adelie 73 73 5## Chinstrap 34 34 0## Gentoo 58 61 4What adornments does the tabyl to right have?
penguins %>% tabyl(species, sex) %>% adorn_percentages(denominator = "col") %>% adorn_totals("row") %>% adorn_pct_formatting(digits = 1) %>% adorn_ns()## species female male NA_## Adelie 44.2% (73) 43.5% (73) 55.6% (5)## Chinstrap 20.6% (34) 20.2% (34) 0.0% (0)## Gentoo 35.2% (58) 36.3% (61) 44.4% (4)## Total 100.0% (165) 100.0% (168) 100.0% (9)- Base R has a
tablefunction, but it is clunkier and the output is not a data frame (or tibble). - See the tabyl vignette for more information, adorn options, & 3-way
tabyls
3 way tabyls are possible
penguins %>% tabyl(species, island, sex)## $female## species Biscoe Dream Torgersen## Adelie 22 27 24## Chinstrap 0 34 0## Gentoo 58 0 0## ## $male## species Biscoe Dream Torgersen## Adelie 22 28 23## Chinstrap 0 34 0## Gentoo 61 0 0## ## $NA_## species Biscoe Dream Torgersen## Adelie 0 1 4## Chinstrap 0 0 0## Gentoo 4 0 0Practice 3
Continue adding code chunks to your Rmd (or, start a new one! But remember to load the libraries and data at the top.)
How many different years are in the data? (Hint: use
tabyl()orn_distinct())Count the number of penguins measured each year.
Calculate the median body mass by each species and sex subgroup. Use
summarize()andgroup_by()to do this.Create a 2x2 table of number of penguins measured in each year by each island.
- Take a break!
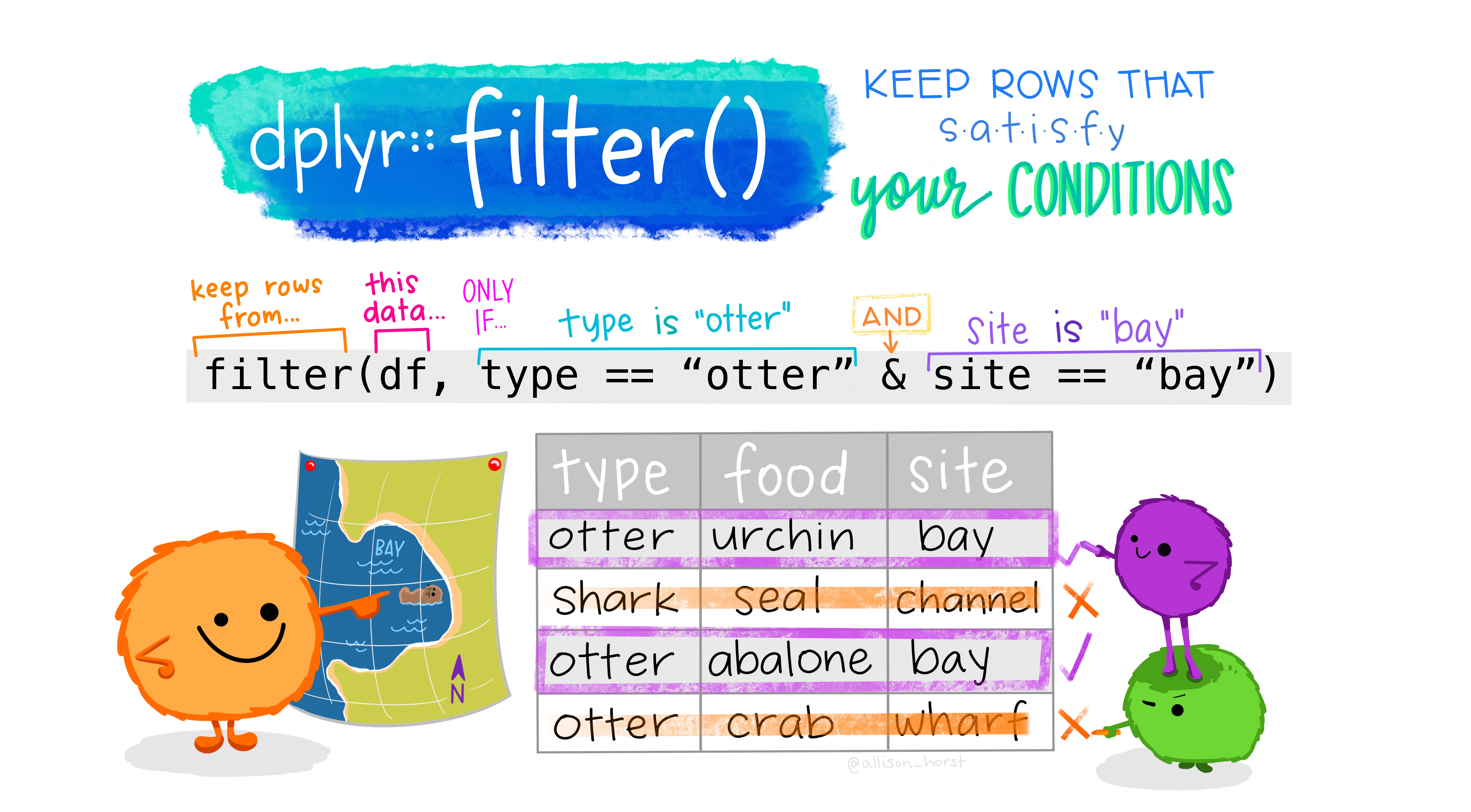
filter() options
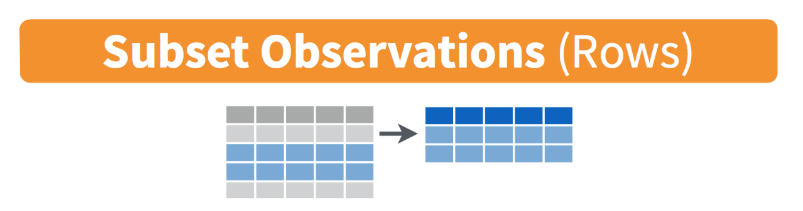
Subset rows of data by specifying conditions within filter()
- math:
>,<,>=,<= - double = for "is equal to":
== &(and)|(or)- != (not equal)
is.na()to filter based on missing values%in%to filter based on group membership!in front negates the statement, as in!is.na(sex)!(species %in% c("Adelie", "Gentoo"))
penguins %>% filter(bill_length_mm > 55)## # A tibble: 5 x 9## id species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g## <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl>## 1 4026 Gentoo Biscoe 59.6 17 230 6050## 2 2415 Gentoo Biscoe 55.9 17 228 5600## 3 4629 Gentoo Biscoe 55.1 16 230 5850## 4 2009 Chinst… Dream 58 17.8 181 3700## 5 4452 Chinst… Dream 55.8 19.8 207 4000## # … with 2 more variables: sex <chr>, year <dbl>filter() practice
What do these commands do? Try them out:
penguins %>% filter(island == "Torgersen")penguins %>% filter(bill_length_mm/bill_depth_mm > 3) # can do mathpenguins %>% filter((body_mass_g < 3000) | (body_mass_g > 6000))# filter on multiple variables:penguins %>% filter(body_mass_g < 3000, bill_depth_mm < 20, sex == "female") penguins %>% filter(body_mass_g < 3000 & bill_depth_mm < 20 & sex == "female") penguins %>% filter(body_mass_g < 3000 | bill_depth_mm < 20 | sex == "female") penguins %>% filter(year == 2008) # note the use of == instead of just =penguins %>% filter(sex == "female")penguins %>% filter(!(species == "Adelie"))penguins %>% filter(species %in% c("Chinstrap", "Gentoo"))penguins %>% filter(is.na(sex))penguins %>% filter(!is.na(sex))select() columns
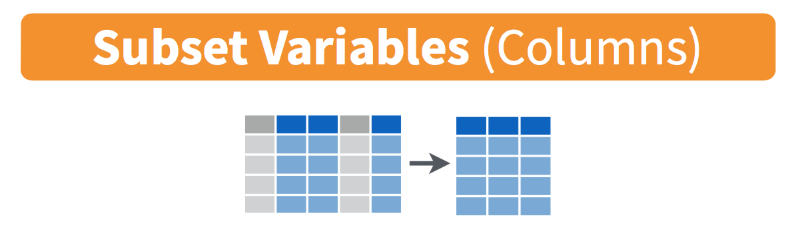
- select columns (variables)
- no quotes needed around variable names
- can be used to rearrange columns
- syntax is flexible and has many options
penguins %>% select(id, island, species, body_mass_g)## # A tibble: 342 x 4## id island species body_mass_g## <dbl> <chr> <chr> <dbl>## 1 1689 Torgersen Adelie 3750## 2 4274 Torgersen Adelie 3800## 3 4539 Torgersen Adelie 3250## 4 2435 Torgersen Adelie 3450## 5 2326 Torgersen Adelie 3650## 6 2637 Torgersen Adelie 3625## 7 4443 Torgersen Adelie 4675## 8 2102 Torgersen Adelie 3475## 9 2975 Torgersen Adelie 4250## 10 3966 Torgersen Adelie 3300## # … with 332 more rowsColumn selection syntax options
There are many ways to select a set of variable names (columns):
var1:var20: all columns fromvar1tovar20- Removing columns
-var1: remove the columnvar1-(var1:var20): remove all columns fromvar1tovar20
- Select by specifying text within column names
contains("mm"),contains("_"): all variable names that contain the specified stringstarts_with("a")orends_with("last"): all variable names that start or end with the specified string
See other examples in the data wrangling cheatsheet.
select() practice
Which columns are selected & in what order using these commands?
First guess and then try them out.
penguins %>% select(id:bill_length_mm)penguins %>% select(where(is.character))penguins %>% select(where(is.numeric))penguins %>% select(-id,-species)penguins %>% select(-(id:island))penguins %>% select(contains("bill"))penguins %>% select(starts_with("s"))penguins %>% select(-contains("mm"))relocate() columns
- change the order of columns in dataset
- default action is to list specified column names first
- no quotes needed around variable names
- similar options as with
select(), plus special ones such as.beforeand.after
penguins %>% relocate(year, body_mass_g)## # A tibble: 342 x 9## year body_mass_g id species island bill_length_mm bill_depth_mm## <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>## 1 2007 3750 1689 Adelie Torge… 39.1 18.7## 2 2007 3800 4274 Adelie Torge… NA 17.4## 3 2007 3250 4539 Adelie Torge… 40.3 18 ## 4 2007 3450 2435 Adelie Torge… 36.7 19.3## 5 2007 3650 2326 Adelie Torge… 39.3 20.6## 6 2007 3625 2637 Adelie Torge… 38.9 17.8## 7 2007 4675 4443 Adelie Torge… NA 19.6## 8 2007 3475 2102 Adelie Torge… 34.1 18.1## 9 2007 4250 2975 Adelie Torge… 42 20.2## 10 2007 3300 3966 Adelie Torge… 37.8 17.1## # … with 332 more rows, and 2 more variables: flipper_length_mm <dbl>,## # sex <chr>relocate() practice
What order are the columns in using these commands?
First guess and then try them out.
penguins %>% relocate(species:bill_length_mm)penguins %>% relocate(where(is.character))penguins %>% relocate(where(is.numeric))penguins %>% relocate(flipper_length_mm,.before = bill_length_mm)penguins %>% relocate(species, .after = island)penguins %>% relocate(species, .after = last_col())Save your modified data
- Use a new variable name and the
<-assignment operator to save a modified data frame - You can save the modified data using the same name, but it will replace the previous dataset
penguins_sub <- penguins %>% select(id:island, sex)penguins_sub## # A tibble: 342 x 4## id species island sex ## <dbl> <chr> <chr> <chr> ## 1 1689 Adelie Torgersen male ## 2 4274 Adelie Torgersen female## 3 4539 Adelie Torgersen female## 4 2435 Adelie Torgersen female## 5 2326 Adelie Torgersen male ## 6 2637 Adelie Torgersen female## 7 4443 Adelie Torgersen male ## 8 2102 Adelie Torgersen <NA> ## 9 2975 Adelie Torgersen <NA> ## 10 3966 Adelie Torgersen <NA> ## # … with 332 more rowsmutate() the data
Use mutate() to add new columns to a tibble
- Many options for how to define new column of data
penguins <- penguins %>% mutate(bill_ratio = bill_length_mm / bill_depth_mm)# use = (not <- or ==) to define new variablepenguins %>% select(bill_ratio, bill_length_mm, bill_depth_mm)## # A tibble: 342 x 3## bill_ratio bill_length_mm bill_depth_mm## <dbl> <dbl> <dbl>## 1 2.09 39.1 18.7## 2 NA NA 17.4## 3 2.24 40.3 18 ## 4 1.90 36.7 19.3## 5 1.91 39.3 20.6## 6 2.19 38.9 17.8## 7 NA NA 19.6## 8 1.88 34.1 18.1## 9 2.08 42 20.2## 10 2.21 37.8 17.1## # … with 332 more rowsmutate() practice
What do the following commands do?
First guess and then try them out.
penguins <- penguins %>% mutate(bill_long = (bill_length_mm > 45))penguins <- penguins %>% mutate(male = (sex == "male"))penguins <- penguins %>% mutate(male2 = 1 * (sex == "male"))rename() columns
rename(new_name = old_name)
Code renames the column, but just prints output without saving the rename:
# This does not save the new namepenguins %>% rename(record = id)## # A tibble: 342 x 10## record species island bill_length_mm bill_depth_mm flipper_length_…## <dbl> <chr> <chr> <dbl> <dbl> <dbl>## 1 1689 Adelie Torge… 39.1 18.7 181## 2 4274 Adelie Torge… NA 17.4 186## 3 4539 Adelie Torge… 40.3 18 195## 4 2435 Adelie Torge… 36.7 19.3 193## 5 2326 Adelie Torge… 39.3 20.6 190## 6 2637 Adelie Torge… 38.9 17.8 181## 7 4443 Adelie Torge… NA 19.6 195## 8 2102 Adelie Torge… 34.1 18.1 193## 9 2975 Adelie Torge… 42 20.2 190## 10 3966 Adelie Torge… 37.8 17.1 186## # … with 332 more rows, and 4 more variables: body_mass_g <dbl>, sex <chr>,## # year <dbl>, bill_ratio <dbl>Code renames the column and overwrites penguins with renamed column:
penguins2 <- penguins %>% rename(record = id)penguins2## # A tibble: 342 x 10## record species island bill_length_mm bill_depth_mm flipper_length_…## <dbl> <chr> <chr> <dbl> <dbl> <dbl>## 1 1689 Adelie Torge… 39.1 18.7 181## 2 4274 Adelie Torge… NA 17.4 186## 3 4539 Adelie Torge… 40.3 18 195## 4 2435 Adelie Torge… 36.7 19.3 193## 5 2326 Adelie Torge… 39.3 20.6 190## 6 2637 Adelie Torge… 38.9 17.8 181## 7 4443 Adelie Torge… NA 19.6 195## 8 2102 Adelie Torge… 34.1 18.1 193## 9 2975 Adelie Torge… 42 20.2 190## 10 3966 Adelie Torge… 37.8 17.1 186## # … with 332 more rows, and 4 more variables: body_mass_g <dbl>, sex <chr>,## # year <dbl>, bill_ratio <dbl>Practice 4
Create a new Rmd or continue in your current Rmd.
Create a dataset for just the Torgersen island penguins that are female.
Restrict the data to just Torgersen female penguins that weigh more than 3500 g.
Restrict the dataset from the previous step to include just the columns with the original body measurements.
Add a column for the difference in the flipper and bill lengths, and call it
flipper_bill_diff.How many rows and columns does your final dataset have?
- Take a break!
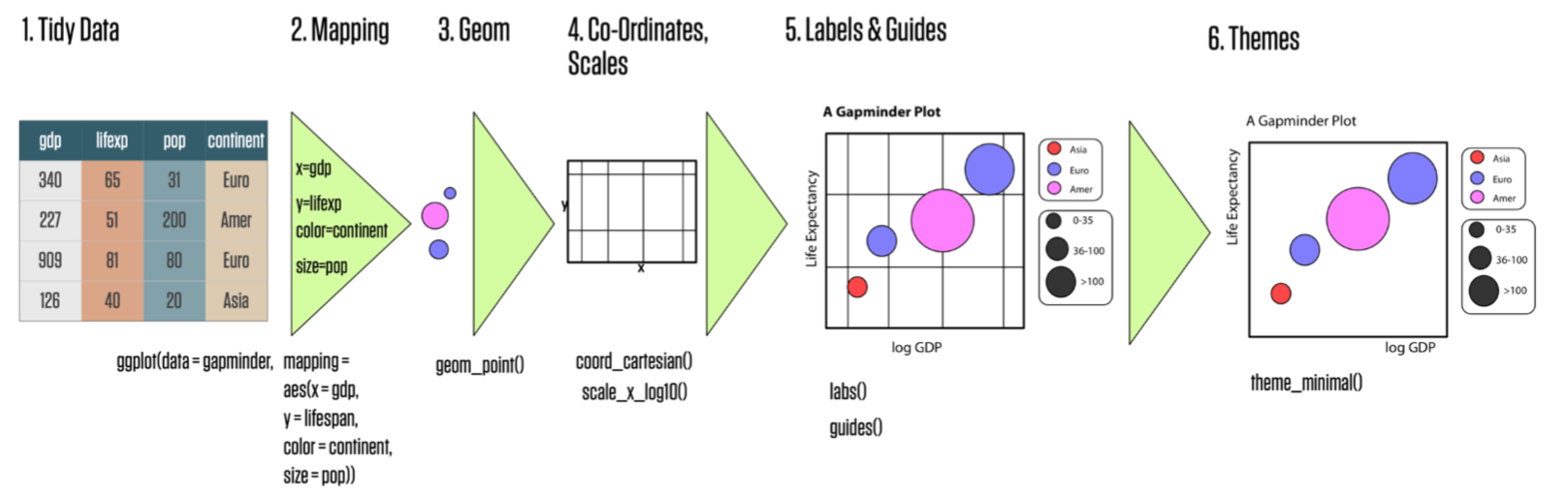
Basics of ggplot
- For a full treatment, watch our BERD workshop "Data Visualization with R and ggplot2"
Back to ggplot code

Ggplot needs tidy data
What are tidy data?
- Each variable forms a column
- Each observation forms a row
- Each value has its own cell
 G. Grolemond & H. Wickham's R for Data Science
G. Grolemond & H. Wickham's R for Data Science
See BERD workshop Data Wrangling Part 1 slides for more info.
Is our data tidy?
Simple plots
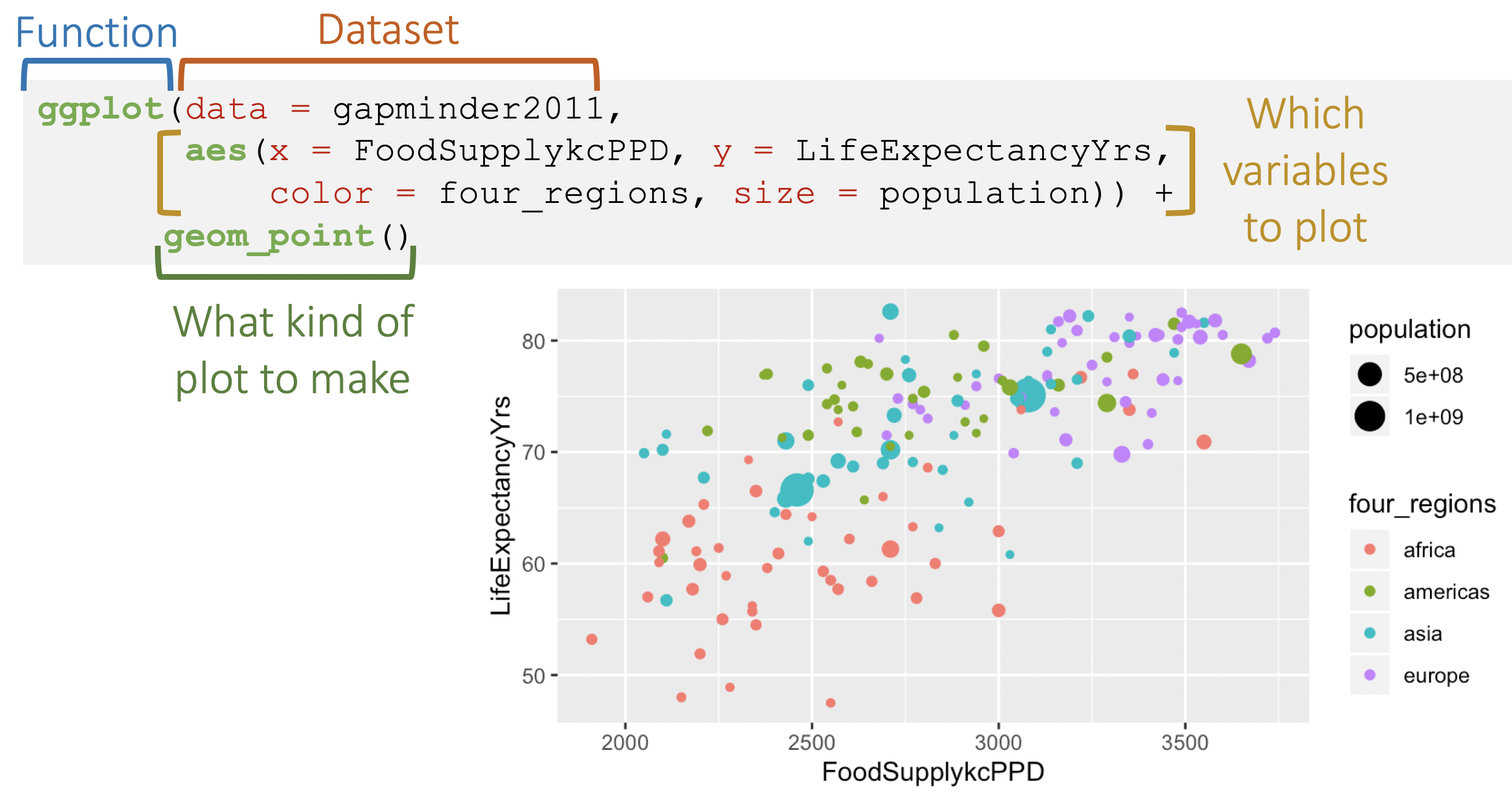
ggplot(data = penguins, aes(x = flipper_length_mm, y = bill_length_mm)) + geom_point()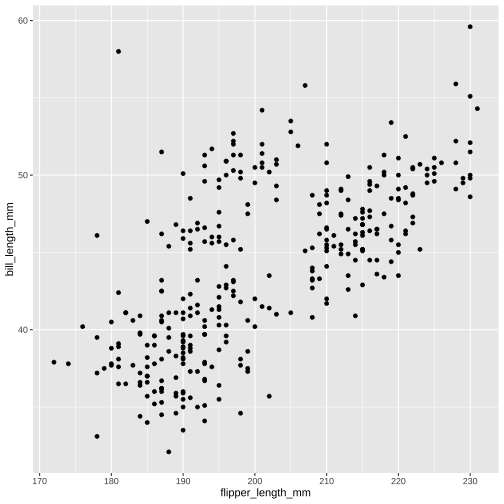
ggplot(data = penguins, aes(x = flipper_length_mm)) + geom_histogram()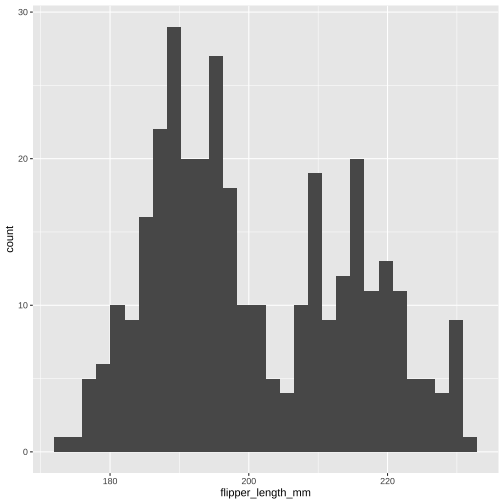
Improved plots - tips
Start with simple, slowly add in additions/colors/etc
You are building a plot layer by layer! ++++++
At the beginning, just copy and paste examples that you want to edit until you understand what each function does
It will take some trial and error!
Watch BERD ggplot video for more instruction, and many customizations
Improved scatterplot
ggplot(data = penguins, aes(x = flipper_length_mm, y = bill_length_mm, color = species)) + geom_point()+ labs( title = "Flipper & bill length", subtitle = "Palmer Station LTER", x = "Flipper length(mm)", y = "Bill length(mm)") + scale_color_viridis_d( name = "Penguin species") + theme_bw()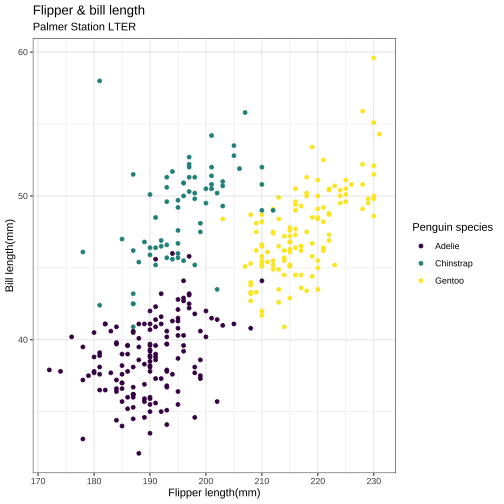
Improved histogram
ggplot(data = penguins, aes(x = flipper_length_mm, fill = species)) + geom_histogram( alpha = 0.5, position = "identity") + labs( title = "Flipper length", x = "Flipper length(mm)", y = "Frequency") + scale_fill_viridis_d( name = "Penguin species") + theme_minimal()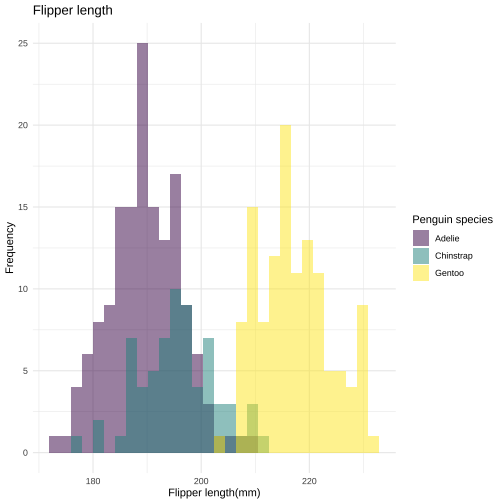
Boxplot + jitter
ggplot(data = penguins, aes(x = species, y = flipper_length_mm)) + geom_boxplot(color="darkgrey", width = 0.3, show.legend = FALSE) + geom_jitter( aes(color = species), alpha = 0.5, show.legend = FALSE, position = position_jitter( width = 0.2, seed = 0)) + scale_color_manual( values = c("darkorange","purple", "cyan4")) + theme_minimal() + labs(x = "Species", y = "Flipper length (mm)")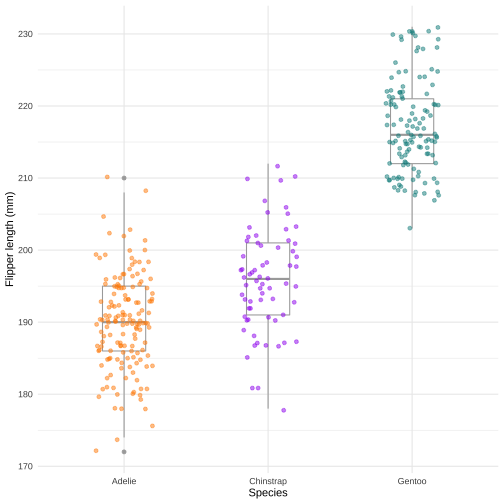
Bar plots - counts
ggplot(data = penguins, aes(x = species, fill = sex)) + geom_bar()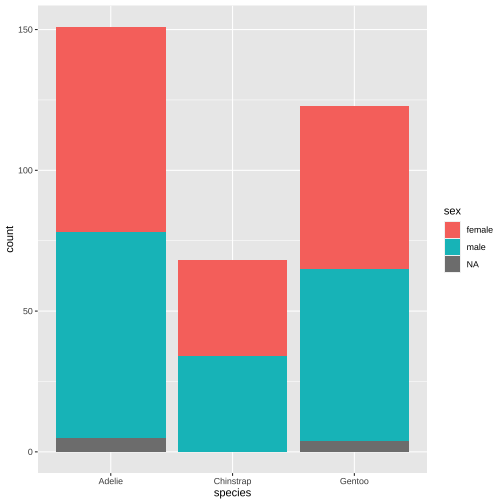
ggplot(data = penguins, aes(x = species, fill = sex)) + geom_bar(position = "dodge")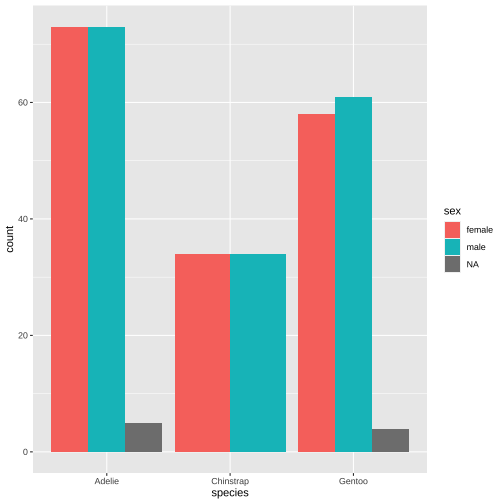
Bar plots - percentages
pct_data <- penguins %>% count(species, sex) %>% # filter(!is.na(sex)) %>% group_by(species) %>% mutate(pct = 100*n/sum(n))pct_data## # A tibble: 8 x 4## # Groups: species [3]## species sex n pct## <chr> <chr> <int> <dbl>## 1 Adelie female 73 48.3 ## 2 Adelie male 73 48.3 ## 3 Adelie <NA> 5 3.31## 4 Chinstrap female 34 50 ## 5 Chinstrap male 34 50 ## 6 Gentoo female 58 47.2 ## 7 Gentoo male 61 49.6 ## 8 Gentoo <NA> 4 3.25ggplot(data = pct_data, aes(x = species, y = pct, fill = sex)) + geom_col()+ ylab("Percent")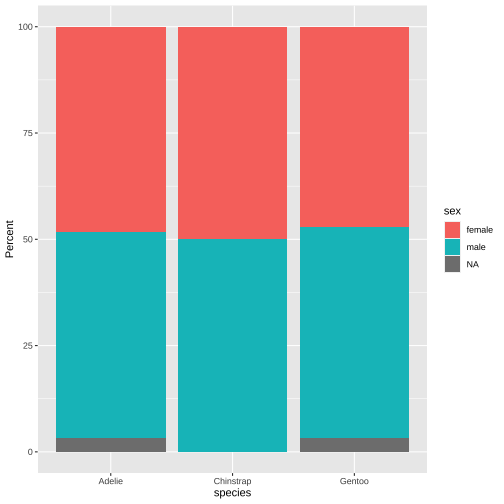
Bar plots - percentages
pct_data <- penguins %>% count(species, sex) %>% # filter(!is.na(sex)) %>% group_by(species) %>% mutate(pct = 100*n/sum(n))pct_data## # A tibble: 8 x 4## # Groups: species [3]## species sex n pct## <chr> <chr> <int> <dbl>## 1 Adelie female 73 48.3 ## 2 Adelie male 73 48.3 ## 3 Adelie <NA> 5 3.31## 4 Chinstrap female 34 50 ## 5 Chinstrap male 34 50 ## 6 Gentoo female 58 47.2 ## 7 Gentoo male 61 49.6 ## 8 Gentoo <NA> 4 3.25ggplot(data = pct_data, aes(x = species, y = pct, fill = sex)) + geom_col(position = "dodge") + ylab("Percent")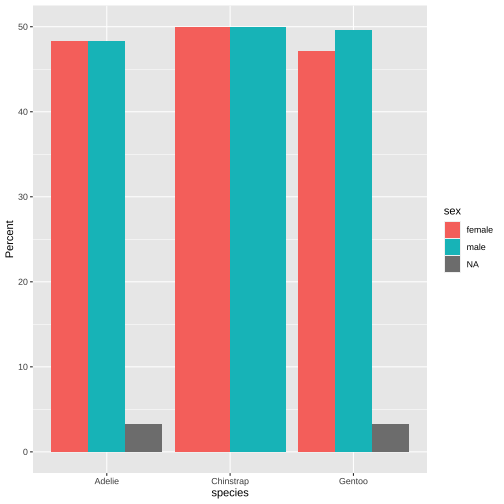
Practice 5
Continue adding code chunks to your Rmd (or, start a new one! But remember to load the libraries and data at the top.)
Make a scatter plot of bill depth vs bill length.
Add
+ geom_smooth(method="lm")to the plot. What is this saying about the association between bill depth and length?Now add
color = speciesto the aestheticaes(). Keepgeom_smooth. How do the associations look now?
Factors for categorical data
factoris a data type that saves character variables as categories (factor levels)Using factor data types are useful for making plots and necessary for some statistical modeling functions
We recommend using commands from the
forcatspackage to work with factor dataSee
forcatscheatsheet andforcatsvignette
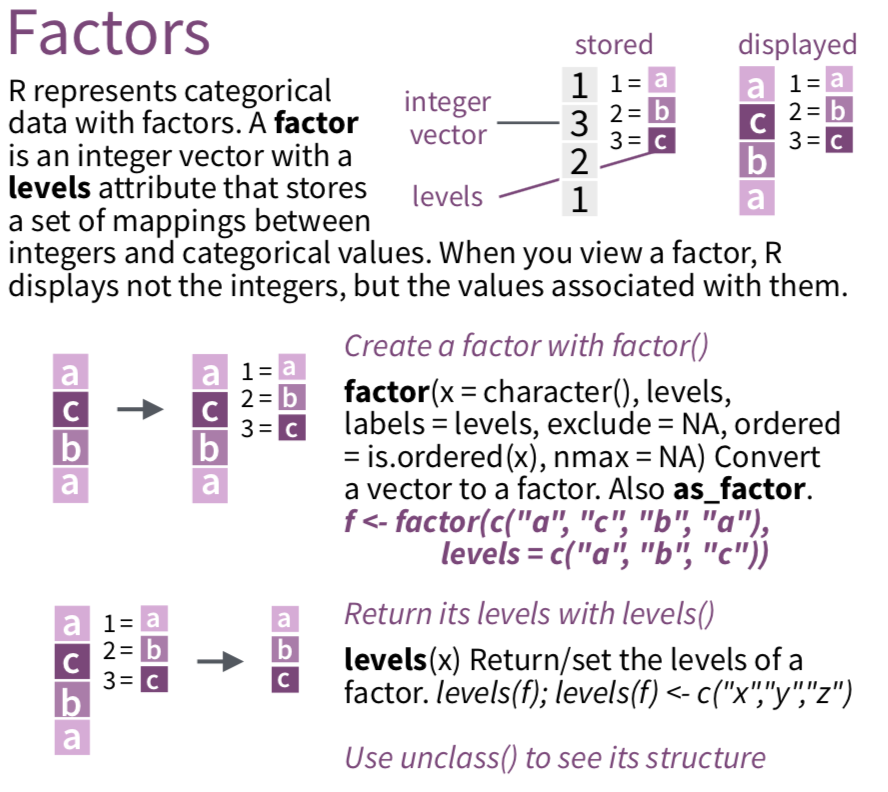
Create a factor variable using factor()
penguins <- penguins %>% mutate(sex_fac = factor(sex))levels(penguins$sex_fac) # factor levels are in alphanumeric order by default## [1] "female" "male"penguins %>% select(sex, sex_fac) %>% summary() # character vs. factor types## sex sex_fac ## Length:342 female:165 ## Class :character male :168 ## Mode :character NA's : 9penguins %>% select(sex, sex_fac) %>% str() # str for structure## tibble [342 × 2] (S3: tbl_df/tbl/data.frame)## $ sex : chr [1:342] "male" "female" "female" "female" ...## $ sex_fac: Factor w/ 2 levels "female","male": 2 1 1 1 2 1 2 NA NA NA ...Specify order of factor levels: fct_relevel()
penguins <- penguins %>% mutate(species_fac = factor(species))summary(penguins$species_fac) # levels are in alphanumeric order by default## Adelie Chinstrap Gentoo ## 151 68 123penguins <- penguins %>% mutate(species_fac = fct_relevel(species_fac, c("Adelie", "Gentoo", "Chinstrap")))summary(penguins$species_fac) # levels are specified order## Adelie Gentoo Chinstrap ## 151 123 68Collapse factor levels
penguins <- penguins %>% mutate(species_fac2 = fct_collapse(species_fac, # collapse levels Adelie = c("Adelie"), Other = c("Gentoo", "Chinstrap")) )penguins %>% select(species_fac, species_fac2) %>% summary()## species_fac species_fac2## Adelie :151 Adelie:151 ## Gentoo :123 Other :191 ## Chinstrap: 68penguins %>% tabyl(species_fac, species_fac2)## species_fac Adelie Other## Adelie 151 0## Gentoo 0 123## Chinstrap 0 68The more you know
Save data frame
- Save .RData file: the standard R format, which is recommended if saving data for future use in R
save(penguins, file = "penguins.RData") # saving mydata within the data folderYou can load .RData files using the load() command:
load("penguins.RData")- Save csv file: comma-separated values
write_csv(penguins, path = "my_penguin_data.csv")How to get help (1/2)
Use ? in front of function name in console. Try this:
How to get help (2/2)
- Use
??(i.e??dplyror??read_csv) for searching all documentation in installed packages (including unloaded packages) - search Stack Overflow #r tag
- googlequestion + rcran or + r (i.e. "make a boxplot rcran" "make a boxplot r")
- google error in quotes (i.e.
"Evaluation error: invalid type (closure) for variable '***'") - search github for your function name (to see examples) or error
- Rstudio community
- twitter #rstats
Resources
- Click on this LONG LIST of resources for learning R
- Watch recordings of our other workshops with all slides/materials at github.com/jminnier/berd_r_courses
Recommended viewing order of BERD workshops:
- Either this workshop, or the older version: Getting Started with R and Rstudio (September 24, 2019, 3 hour version)
- no tidyverse, just base R introduction; The new version we are doing now is hopefully better!
- Data Wrangling in R, Part 1A and 1B (April 18, 2019, 2 hours)
- more data wrangling, tidy data, removing missing data, arranging data
- Data Wrangling in R, Part 2 (April 25, 2019, 2 hours)
- even more data wrangling, adding columns, summarizing, joining/merging two or more data sets together, reshaping data from wide to long format or vice versa, more methods for dealing with NAs, working with dates and factors, cleaning up messy column names
- Data Visualization with R and ggplot2 (May 20, 2020, 2.5 hours)
- additional ggplot examples, many more ways to customize your ggplots!
Other Resources
Getting started:
- Again, check out this LONG LIST of resources for learning R
- R Bootcamp - by Ted Laderas and Jessica Minnier
- Rstudio primers
- Teacup Giraffes for learning stats & R
Basic help with installation and using Rstudio
- RStudio IDE Cheatsheet
- Install R/RStudio help video
- Basic Basics
Some of this is drawn from materials in online books/lessons:
- Intro to R/RStudio by Emma Rand
- Modern Dive - An Introduction to Statistical and Data Sciences via R by Chester Ismay & Albert Kim
- R for Data Science - Hadley Wickham & Garrett Grolemund
- Cookbook for R by Winston Chang
Local resources
- OHSU's BioData club + active slack channel
- Portland's R user meetup group + active slack channel
- R-ladies PDX meetup group
- Cascadia R Conf - click on Years for old videos
Contact info:
Jessica Minnier: minnier@ohsu.edu
Meike Niederhausen: niederha@ohsu.edu
This workshop info:
- Code for these slides on github: jminnier/berd_r_courses
- Link to html: part 1 & part 2
- all the R code in an R script: part 1 & part 2
- answers to practice problems can be found here: part 1 & part 2