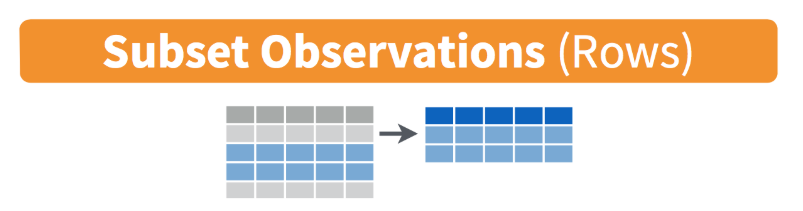
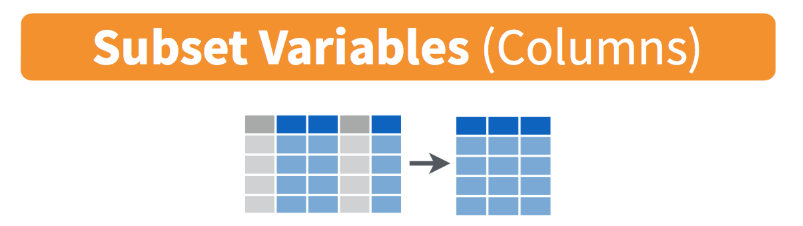
Data Wrangling in R with the Tidyverse (Part 1)
Jessica Minnier, PhD & Meike Niederhausen, PhD
OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop
2019/04/18 (Part 1) & 2019/04/25 (Part 2)
and again! 2019/05/16 (Part 1) & 2019/05/23 (Part 2)
slides: bit.ly/berd_tidy1
pdf: bit.ly/berd_tidy1_pdf
Load files for today's workshop
- Open the slides of this workshop: bit.ly/berd_tidy1
- Open the pre-workshop homework
- Follow steps 1-5: Download zip folder, open
berd_tidyverse_project.Rproj - Open a new R script and run the commands to the right
# install.packages("tidyverse")library(tidyverse)library(lubridate)demo_data <- read_csv("data/yrbss_demo.csv")

Learning objectives
Part 1:
- What is data wrangling?
- A few good practices in R/RStudio
- What is tidy data?
- What is tidyverse?
- Manipulate data
Part 2:
- Reshaping (long/wide format) data
- Join/merge data sets
- Data cleaning, including examples for dealing with:
- Missing data
- Strings/character vectors
- Factors/categorical variables
- Dates
What is data wrangling?
- "data janitor work"
- importing data
- cleaning data
- changing shape of data
- fixing errors and poorly formatted data elements
- transforming columns and rows
- filtering, subsetting
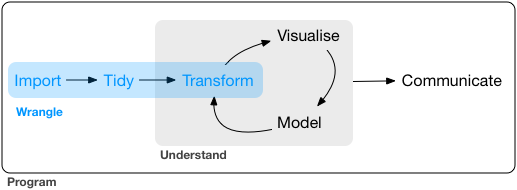
G. Grolemond & H. Wickham's R for Data Science
Good practices in RStudio
Use projects (read this)
- Create an RStudio project for each data analysis project
A project is associated with a directory folder
- Keep data files there
- Keep scripts there; edit them, run them in bits or as a whole
- Save your outputs (plots and cleaned data) there
Only use relative paths, never absolute paths
- relative (good):
read_csv("data/mydata.csv") - absolute (bad):
read_csv("/home/yourname/Documents/stuff/mydata.csv")
- relative (good):
Advantages of using projects
- standardize file paths
- keep everything together
- a whole folder can be shared and run on another computer
Useful keyboard shortcuts
| action | mac | windows/linux |
|---|---|---|
| run code in script | cmd + enter | ctrl + enter |
<- |
option + - | alt + - |
%>% (covered later) |
cmd + shift + m | ctrl + shift + m |
Try typing (with shortcut) and running
y <- 5yNow, in the console, press the up arrow.
Others: (see full list)
| action | mac | windows/linux |
|---|---|---|
| interrupt currently executing command | esc | esc |
| in console, go to previously run code | up/down | up/down |
| keyboard shortcut help | option + shift + k | alt + shift + k |
Data frames vs. tibbles
Previously we learned about data frames
data.frame(name = c("Sarah","Ana","Jose"), rank = 1:3, age = c(35.5, 25, 58), city = c(NA,"New York","LA")) name rank age city1 Sarah 1 35.5 <NA>2 Ana 2 25.0 New York3 Jose 3 58.0 LAA tibble is a data frame but with perks
tibble(name = c("Sarah","Ana","Jose"), rank = 1:3, age = c(35.5, 25, 58), city = c(NA,"New York","LA"))# A tibble: 3 x 4 name rank age city <chr> <int> <dbl> <chr> 1 Sarah 1 35.5 <NA> 2 Ana 2 25 New York3 Jose 3 58 LAHow are these two datasets different?
Import data as a data frame (try this)
Base R functions import data as data frames (read.csv, read.table, etc)
mydata_df <- read.csv("data/small_data.csv")mydata_df id age sex grade race41 335340 17 years old Female 10th White2 638618 16 years old Female 9th <NA>3 922382 14 years old Male 9th White4 923122 15 years old Male 9th White5 923963 15 years old Male 10th Black or African American6 925603 16 years old Male 10th All other races7 933724 16 years old Female 10th All other races8 935435 17 years old Female 12th All other races9 1096564 15 years old Male 10th All other races10 1108114 17 years old Female 9th Black or African American11 1306150 16 years old Male 10th Hispanic/Latino12 1307481 17 years old Male 12th Hispanic/Latino13 1307872 17 years old Male 11th Hispanic/Latino14 1311617 15 years old Female 10th Hispanic/Latino15 1313153 16 years old Female 11th Hispanic/Latino16 1313291 16 years old Female 11th White17 1313477 16 years old Female 10th All other races18 1315121 17 years old Female 11th <NA>19 1315850 17 years old Female 12th Hispanic/Latino20 1316123 18 years old or older Female 12th Black or African American bmi weight_kg text_while_driving_30d smoked_ever1 27.5671 66.23 <NA> <NA>2 29.3495 84.82 <NA> Yes3 18.1827 57.61 <NA> Yes4 21.3754 60.33 <NA> Yes5 19.5988 63.50 <NA> No6 22.1910 70.31 <NA> No7 20.9913 45.36 <NA> Yes8 17.4814 43.09 <NA> No9 22.4593 79.38 <NA> <NA>10 26.5781 68.04 <NA> No11 21.1874 67.13 0 days <NA>12 19.4637 56.25 1 or 2 days No13 20.6121 61.69 1 or 2 days No14 27.4648 70.31 0 days No15 26.5781 68.04 0 days No16 24.8047 63.50 3 to 5 days No17 25.0318 76.66 0 days No18 22.2687 54.89 I did not drive the past 30 days Yes19 19.4922 49.90 0 days <NA>20 27.4894 74.84 All 30 days Yes bullied_past_12mo height_m1 NA 1.5500002 NA 1.6999993 FALSE 1.7799994 FALSE 1.6800015 TRUE 1.7999986 TRUE 1.7800007 TRUE 1.4699988 FALSE 1.5700029 TRUE 1.87999810 FALSE 1.60000111 FALSE 1.77999812 FALSE 1.69999913 FALSE 1.73000114 TRUE 1.60000115 TRUE 1.60000116 FALSE 1.60000017 TRUE 1.75000118 FALSE 1.56999819 FALSE 1.59999920 FALSE 1.650001Import data as a tibble (try this)
tidyverse functions import data as tibbles (read_csv, read_excel(), etc)
mydata_tib <- read_csv("data/small_data.csv")mydata_tib# A tibble: 20 x 11 id age sex grade race4 bmi weight_kg text_while_driv… <dbl> <chr> <chr> <chr> <chr> <dbl> <dbl> <chr> 1 3.35e5 17 y… Fema… 10th White 27.6 66.2 <NA> 2 6.39e5 16 y… Fema… 9th <NA> 29.3 84.8 <NA> 3 9.22e5 14 y… Male 9th White 18.2 57.6 <NA> 4 9.23e5 15 y… Male 9th White 21.4 60.3 <NA> 5 9.24e5 15 y… Male 10th Blac… 19.6 63.5 <NA> 6 9.26e5 16 y… Male 10th All … 22.2 70.3 <NA> 7 9.34e5 16 y… Fema… 10th All … 21.0 45.4 <NA> 8 9.35e5 17 y… Fema… 12th All … 17.5 43.1 <NA> 9 1.10e6 15 y… Male 10th All … 22.5 79.4 <NA> 10 1.11e6 17 y… Fema… 9th Blac… 26.6 68.0 <NA> 11 1.31e6 16 y… Male 10th Hisp… 21.2 67.1 0 days 12 1.31e6 17 y… Male 12th Hisp… 19.5 56.2 1 or 2 days 13 1.31e6 17 y… Male 11th Hisp… 20.6 61.7 1 or 2 days 14 1.31e6 15 y… Fema… 10th Hisp… 27.5 70.3 0 days 15 1.31e6 16 y… Fema… 11th Hisp… 26.6 68.0 0 days 16 1.31e6 16 y… Fema… 11th White 24.8 63.5 3 to 5 days 17 1.31e6 16 y… Fema… 10th All … 25.0 76.7 0 days 18 1.32e6 17 y… Fema… 11th <NA> 22.3 54.9 I did not drive…19 1.32e6 17 y… Fema… 12th Hisp… 19.5 49.9 0 days 20 1.32e6 18 y… Fema… 12th Blac… 27.5 74.8 All 30 days # … with 3 more variables: smoked_ever <chr>, bullied_past_12mo <lgl>,# height_m <dbl>Compare & contrast data frame and tibble
Run the code below
data frame
glimpse(mydata_df)str(mydata_df) # How are glimpse() and str() different?head(mydata_df)summary(mydata_df)class(mydata_df) # What information does class() give?tibble
glimpse(mydata_tib)str(mydata_tib) head(mydata_tib)summary(mydata_tib)class(mydata_tib)Tibble perks
Viewing tibbles:
- variable types are given (character, factor, double, integer, boolean, date)
- number of rows & columns shown are limited for easier viewing
Other perks:
- tibbles can typically be used anywhere a
data.frameis needed read_*()functions don't read character columns as factors (no surprises)
What are tidy data?
- Each variable forms a column
- Each observation forms a row
- Each value has its own cell
 G. Grolemond & H. Wickham's R for Data Science
G. Grolemond & H. Wickham's R for Data Science
Untidy data: example 1
untidy_data <- tibble( name = c("Ana","Bob","Cara"), meds = c("advil 600mg 2xday","tylenol 650mg 4xday", "advil 200mg 3xday"))untidy_data# A tibble: 3 x 2 name meds <chr> <chr> 1 Ana advil 600mg 2xday 2 Bob tylenol 650mg 4xday3 Cara advil 200mg 3xdayTidy data: example 1
You will learn how to do this!
untidy_data %>% separate(col = meds, into = c("med_name","dose_mg","times_per_day"), sep=" ") %>% mutate(times_per_day = as.numeric(str_remove(times_per_day, "xday")), dose_mg = as.numeric(str_remove(dose_mg, "mg")))# A tibble: 3 x 4 name med_name dose_mg times_per_day <chr> <chr> <dbl> <dbl>1 Ana advil 600 22 Bob tylenol 650 43 Cara advil 200 3Untidy data: example 2
untidy_data2 <- tibble( name = c("Ana","Bob","Cara"), wt_07_01_2018 = c(100, 150, 140), wt_08_01_2018 = c(104, 155, 138), wt_09_01_2018 = c(NA, 160, 142))untidy_data2# A tibble: 3 x 4 name wt_07_01_2018 wt_08_01_2018 wt_09_01_2018 <chr> <dbl> <dbl> <dbl>1 Ana 100 104 NA2 Bob 150 155 1603 Cara 140 138 142Tidy data: example 2
You will learn how to do this!
untidy_data2 %>% gather(key = "date", value = "weight", -name) %>% mutate(date = str_remove(date,"wt_"), date = dmy(date)) # dmy() is a function in the lubridate package# A tibble: 9 x 3 name date weight <chr> <date> <dbl>1 Ana 2018-01-07 1002 Bob 2018-01-07 1503 Cara 2018-01-07 1404 Ana 2018-01-08 1045 Bob 2018-01-08 1556 Cara 2018-01-08 1387 Ana 2018-01-09 NA8 Bob 2018-01-09 1609 Cara 2018-01-09 142Tools for tidying data
tidyversefunctionstidyverseis a suite of packages that implementtidymethods for data importing, cleaning, and wrangling- load the
tidyversepackages by running the codelibrary(tidyverse)- see pre-workshop homework for code to install
tidyverse
- see pre-workshop homework for code to install
Functions to easily work with rows and columns, such as
- subset rows/columns
- add new rows/columns
- split apart or unite columns
- join together different data sets (part 2)
- make data long or wide (part 2)
Often many steps to tidy data
- string together commands to be performed sequentially
- do this using pipes
%>%
How to use the pipe %>%
The pipe operator %>% strings together commands to be performed sequentially
mydata_tib %>% head(n=3) # prounounce %>% as "then"# A tibble: 3 x 11 id age sex grade race4 bmi weight_kg text_while_driv… <dbl> <chr> <chr> <chr> <chr> <dbl> <dbl> <chr> 1 335340 17 y… Fema… 10th White 27.6 66.2 <NA> 2 638618 16 y… Fema… 9th <NA> 29.3 84.8 <NA> 3 922382 14 y… Male 9th White 18.2 57.6 <NA> # … with 3 more variables: smoked_ever <chr>, bullied_past_12mo <lgl>,# height_m <dbl>- Always first list the tibble that the commands are being applied to
- Can use multiple pipes to run multiple commands in sequence
- What does the following code do?
mydata_tib %>% head(n=3) %>% summary()About the data
Data from the CDC's Youth Risk Behavior Surveillance System (YRBSS)
- complex survey data
- national school-based survey
- conducted by CDC and state, territorial, and local education and health agencies and tribal governments
- monitors six categories of health-related behaviors
- that contribute to the leading causes of death and disability among youth and adults
- including alcohol & drug use, unhealthy & dangerous behaviors, sexuality, and physical activity
- see Questionnaires
the data in
yrbss_demo.csvare a subset of data in the R packageyrbss, which includes YRBSS from 1991-2013Look at your Environment tab to make sure
demo_datais already loaded
demo_data <- read_csv("data/yrbss_demo.csv")filter() ∼ rows
filter data based on rows
- math:
>,<,>=,<= - double = for "is equal to":
== &(and)|(or)- != (not equal)
is.na()to filter based on missing values%in%to filter based on group membership!in front negates the statement, as in!is.na(age)!(grade %in% c("9th","10th"))
demo_data %>% filter(bmi > 20)# A tibble: 10,375 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> 1 333862 17 years o… Fema… 12th White White 20.2 57.2 2 1095530 15 years o… Male 10th Black or Af… Black or Af… 28.0 85.7 3 1303997 14 years o… Male 9th All other r… Multiple - … 24.5 66.7 4 926649 16 years o… Male 11th All other r… Asian 20.5 70.3 5 506337 18 years o… Male 12th Hispanic/La… Hispanic/La… 33.1 123. 6 1307180 16 years o… Male 10th Hispanic/La… Hispanic/La… 21.8 66.7 7 1312128 15 years o… Fema… 10th White White 22.0 65.8 8 770177 16 years o… Fema… 10th White White 32.4 86.2 9 938291 18 years o… Fema… 12th White White 21.7 64.910 1306691 16 years o… Male 11th White White 28.3 102. # … with 10,365 more rowsCompare to base R
- Bracket method: need to repeat tibble name
- Need to use
$ - Very nested and confusing to read
- Keeps
NAs
demo_data[demo_data$grade=="9th",]# A tibble: 5,625 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> 1 1303997 14 years… Male 9th All other… Multiple - Non… 24.5 66.7 2 261619 17 years… Male 9th All other… <NA> NA NA 3 1096939 15 years… Male 9th <NA> <NA> 17.1 45.4 4 180968 15 years… Male 9th White White NA NA 5 924270 15 years… Male 9th All other… Asian 30.7 81.6 6 330828 15 years… Female 9th Hispanic/… Hispanic/Latino 20.4 52.2 7 1311252 15 years… Female 9th Hispanic/… Hispanic/Latino NA NA 8 36853 14 years… Female 9th All other… <NA> NA NA 9 1310689 14 years… Female 9th Hispanic/… Hispanic/Latino 22.5 55.310 1310726 14 years… Female 9th All other… Asian 30.7 81.6# … with 5,615 more rows- Pipe method: list tibble name once
- No
$needed since uses "non-standard evaluation":filter()knowsgradeis a column indemo_data - Removes
NAs
demo_data %>% filter(grade=="9th")# A tibble: 5,219 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> 1 1303997 14 years… Male 9th All other… Multiple - Non… 24.5 66.7 2 261619 17 years… Male 9th All other… <NA> NA NA 3 1096939 15 years… Male 9th <NA> <NA> 17.1 45.4 4 180968 15 years… Male 9th White White NA NA 5 924270 15 years… Male 9th All other… Asian 30.7 81.6 6 330828 15 years… Female 9th Hispanic/… Hispanic/Latino 20.4 52.2 7 1311252 15 years… Female 9th Hispanic/… Hispanic/Latino NA NA 8 36853 14 years… Female 9th All other… <NA> NA NA 9 1310689 14 years… Female 9th Hispanic/… Hispanic/Latino 22.5 55.310 1310726 14 years… Female 9th All other… Asian 30.7 81.6# … with 5,209 more rowsfilter() practice
What do these commands do? Try them out:
demo_data %>% filter(bmi < 5)demo_data %>% filter(bmi/stweight < 0.5) # can do mathdemo_data %>% filter((bmi < 15) | (bmi > 50))demo_data %>% filter(bmi < 20, stweight < 50, sex == "Male") # filter on multiple variablesdemo_data %>% filter(record == 506901) # note the use of == instead of just =demo_data %>% filter(sex == "Female")demo_data %>% filter(!(grade == "9th"))demo_data %>% filter(grade %in% c("10th", "11th"))demo_data %>% filter(is.na(bmi))demo_data %>% filter(!is.na(bmi))select() ∼ columns
- select columns (variables)
- no quotes needed around variable names
- can be used to rearrange columns
- uses special syntax that is flexible and has many options
demo_data %>% select(record, grade)# A tibble: 20,000 x 2 record grade <dbl> <chr> 1 931897 10th 2 333862 12th 3 36253 11th 4 1095530 10th 5 1303997 9th 6 261619 9th 7 926649 11th 8 1309082 12th 9 506337 12th 10 180494 10th # … with 19,990 more rowsCompare to base R
- Need brackets
- Need quotes around column names
demo_data[, c("record","age","sex")]# A tibble: 20,000 x 3 record age sex <dbl> <chr> <chr> 1 931897 15 years old Female 2 333862 17 years old Female 3 36253 18 years old or older Male 4 1095530 15 years old Male 5 1303997 14 years old Male 6 261619 17 years old Male 7 926649 16 years old Male 8 1309082 17 years old Male 9 506337 18 years old or older Male 10 180494 14 years old Male # … with 19,990 more rows- No quotes needed and easier to read.
- More flexible, either of following work:
demo_data %>% select(record, age, sex)demo_data %>% select(record:sex)# A tibble: 20,000 x 3 record age sex <dbl> <chr> <chr> 1 931897 15 years old Female 2 333862 17 years old Female 3 36253 18 years old or older Male 4 1095530 15 years old Male 5 1303997 14 years old Male 6 261619 17 years old Male 7 926649 16 years old Male 8 1309082 17 years old Male 9 506337 18 years old or older Male 10 180494 14 years old Male # … with 19,990 more rows# A tibble: 20,000 x 3 record age sex <dbl> <chr> <chr> 1 931897 15 years old Female 2 333862 17 years old Female 3 36253 18 years old or older Male 4 1095530 15 years old Male 5 1303997 14 years old Male 6 261619 17 years old Male 7 926649 16 years old Male 8 1309082 17 years old Male 9 506337 18 years old or older Male 10 180494 14 years old Male # … with 19,990 more rowsColumn selection syntax options
There are many ways to select a set of variable names (columns):
var1:var20: all columns fromvar1tovar20one_of(c("a", "b", "c")): all columns with names in the specified character vector of names- Removing columns
-var1: remove the columnvar1-(var1:var20): remove all columns fromvar1tovar20
- Select using text within column names
contains("date"),contains("_"): all variable names that contain the specified stringstarts_with("a")orends_with("last"): all variable names that start or end with the specificed string
- Rearranging columns
- use
everything()to select all columns not already named - example:
select(var1, var20, everything())moves the columnvar20to the second position
- use
See other examples in the data wrangling cheatsheet.
select() practice
Which columns are selected & in what order using these commands?
First guess and then try them out.
demo_data %>% select(record:sex)demo_data %>% select(one_of(c("age","stweight")))demo_data %>% select(-grade,-sex)demo_data %>% select(-(record:sex))demo_data %>% select(contains("race"))demo_data %>% select(starts_with("r"))demo_data %>% select(-contains("r"))demo_data %>% select(record, race4, race7, everything())rename() ∼ columns
- renames column variables
demo_data %>% rename(id = record) # order: new_name = old_name# A tibble: 20,000 x 8 id age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> 1 931897 15 years o… Fema… 10th White White 17.2 54.4 2 333862 17 years o… Fema… 12th White White 20.2 57.2 3 36253 18 years o… Male 11th Hispanic/La… Hispanic/La… NA NA 4 1095530 15 years o… Male 10th Black or Af… Black or Af… 28.0 85.7 5 1303997 14 years o… Male 9th All other r… Multiple - … 24.5 66.7 6 261619 17 years o… Male 9th All other r… <NA> NA NA 7 926649 16 years o… Male 11th All other r… Asian 20.5 70.3 8 1309082 17 years o… Male 12th White White 19.3 59.0 9 506337 18 years o… Male 12th Hispanic/La… Hispanic/La… 33.1 123. 10 180494 14 years o… Male 10th Black or Af… Black or Af… NA NA # … with 19,990 more rowsPractice
# Remember: to save output into the same tibble you would use <-newdata <- newdata %>% select(-record)# Useful to see what categories are availabledemo_data %>% janitor::tabyl(race7)Do the following data wrangling steps in order so that the output from the previous step is the input for the next step.
Save the results in each step as newdata.
Import
demo_data.csvin thedatafolder if you haven't already done so.Filter
newdatato only keep "Asian" or "Native Hawaiian/other PI" subjects that are in the 9th grade, and save again asnewdata.Filter
newdatato remove subjects younger than 13, and save asnewdata.Remove the column
race4, and save asnewdata.How many rows does the resulting
newdatahave? How many columns?
mutate()
Use mutate() to add new columns to a tibble
- many options in how to define new column of data
newdata <- demo_data %>% mutate(height_m = sqrt(stweight / bmi)) # use = (not <- or ==) to define new variablenewdata %>% select(record, bmi, stweight)# A tibble: 20,000 x 3 record bmi stweight <dbl> <dbl> <dbl> 1 931897 17.2 54.4 2 333862 20.2 57.2 3 36253 NA NA 4 1095530 28.0 85.7 5 1303997 24.5 66.7 6 261619 NA NA 7 926649 20.5 70.3 8 1309082 19.3 59.0 9 506337 33.1 123. 10 180494 NA NA # … with 19,990 more rowsmutate() practice
What do the following commands do?
First guess and then try them out.
demo_data %>% mutate(bmi_high = (bmi > 30))demo_data %>% mutate(male = (sex == "Male"))demo_data %>% mutate(male = 1 * (sex == "Male"))demo_data %>% mutate(grade_num = as.numeric(str_remove(grade, "th")))case_when() with mutate()
Use case_when() to create multi-valued variables that depend on an existing column
- Example: create BMI groups based off of the
bmivariable
demo_data2 <- demo_data %>% mutate( bmi_group = case_when( bmi < 18.5 ~ "underweight", # condition ~ new_value bmi >= 18.5 & bmi <= 24.9 ~ "normal", bmi > 24.9 & bmi <= 29.9 ~ "overweight", bmi > 29.9 ~ "obese") )demo_data2 %>% select(bmi, bmi_group) %>% head()# A tibble: 6 x 2 bmi bmi_group <dbl> <chr> 1 17.2 underweight2 20.2 normal 3 NA <NA> 4 28.0 overweight 5 24.5 normal 6 NA <NA>separate() and unite()
separate(): one column to many
- when one column has multiple types of information
- removes original column by default
demo_data %>% separate(age,c("a","y","o","w","w2"), sep = " ") %>% select(a:w2)# A tibble: 20,000 x 5 a y o w w2 <chr> <chr> <chr> <chr> <chr> 1 15 years old <NA> <NA> 2 17 years old <NA> <NA> 3 18 years old or older 4 15 years old <NA> <NA> 5 14 years old <NA> <NA> 6 17 years old <NA> <NA> 7 16 years old <NA> <NA> 8 17 years old <NA> <NA> 9 18 years old or older10 14 years old <NA> <NA> # … with 19,990 more rowsunite(): many columns to one
- paste columns together using a separator
- removes original columns by default
demo_data %>% unite("sexgr", sex, grade, sep=":") %>% select(sexgr)# A tibble: 20,000 x 1 sexgr <chr> 1 Female:10th 2 Female:12th 3 Male:11th 4 Male:10th 5 Male:9th 6 Male:9th 7 Male:11th 8 Male:12th 9 Male:12th 10 Male:10th # … with 19,990 more rowsseparate() and unite() practice
What do the following commands do?
First guess and then try them out.
demo_data %>% separate(age, c("agenum","yrs"), sep = " ")demo_data %>% separate(age, c("agenum","yrs"), sep = " ", remove = FALSE)demo_data %>% separate(grade, c("grade_n"), sep = "th")demo_data %>% separate(grade, c("grade_n"), sep = "t")demo_data %>% separate(race4, c("race4_1", "race4_2"), sep = "/")demo_data %>% unite("sex_grade", sex, grade, sep = "::::")demo_data %>% unite("sex_grade", sex, grade) # what is the default `sep` for unite?demo_data %>% unite("race", race4, race7) # what happens to NA values?More commands to filter rows
Remove rows with missing data
na.omit removes all rows with any missing (NA) values in any column
demo_data %>% na.omit()# A tibble: 12,897 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> 1 931897 15 years o… Fema… 10th White White 17.2 54.4 2 333862 17 years o… Fema… 12th White White 20.2 57.2 3 1095530 15 years o… Male 10th Black or Af… Black or Af… 28.0 85.7 4 1303997 14 years o… Male 9th All other r… Multiple - … 24.5 66.7 5 926649 16 years o… Male 11th All other r… Asian 20.5 70.3 6 1309082 17 years o… Male 12th White White 19.3 59.0 7 506337 18 years o… Male 12th Hispanic/La… Hispanic/La… 33.1 123. 8 1307180 16 years o… Male 10th Hispanic/La… Hispanic/La… 21.8 66.7 9 1312128 15 years o… Fema… 10th White White 22.0 65.810 770177 16 years o… Fema… 10th White White 32.4 86.2# … with 12,887 more rowsWe will discuss dealing with missing data more in part 2
Remove rows with duplicated data
distinct() removes rows that are duplicates of other rows
data_dups <- tibble( name = c("Ana","Bob","Cara", "Ana"), race = c("Hispanic","Other", "White", "Hispanic"))data_dups# A tibble: 4 x 2 name race <chr> <chr> 1 Ana Hispanic2 Bob Other 3 Cara White 4 Ana Hispanicdata_dups %>% distinct()# A tibble: 3 x 2 name race <chr> <chr> 1 Ana Hispanic2 Bob Other 3 Cara WhiteOrder rows: arrange()
Use arrange() to order the rows by the values in specified columns
demo_data %>% arrange(bmi, stweight) %>% head(n=3)# A tibble: 3 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>1 635432 13 years… Female 9th Hispanic/Lat… Hispanic/Lat… 13.2 27.72 501608 15 years… Male 9th All other ra… Asian 13.2 47.63 1097740 16 years… Male 9th Black or Afr… Black or Afr… 13.3 45.4demo_data %>% arrange(desc(bmi), stweight) %>% head(n=3)# A tibble: 3 x 8 record age sex grade race4 race7 bmi stweight <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>1 324452 16 years old Male 11th Black or Af… Black or Af… 53.9 91.22 1310082 18 years ol… Male 11th Black or Af… Black or Af… 53.5 160. 3 328160 18 years ol… Male <NA> Black or Af… Black or Af… 53.4 128.Practice
Do the following data wrangling steps in order so that the output from the previous step is the input for the next step.
Save the results in each step as newdata.
Import
demo_data.csvin thedatafolder if you haven't already done so.Create a variable called
grade_numthat has the numeric grade number (useas.numeric()).Filter the data to keep only students in grade 11 or higher.
Filter out rows when
bmiisNA.Create a binary variable called
bmi_normalthat is equal to 1 whenbmiis between 18.5 to 24.9 and 0 when it is outside that range.Arrange by
grade_numfrom highest to lowestSave all output to
newdata.
Advanced column commands
Mutating multiple columns at once: mutate_*
- variants of
mutate()that are useful for mutating multiple columns at oncemutate_at(),mutate_if(),mutate_all(), etc.
- which columns get mutated depends on a predicate, can be:
- a function that returns TRUE/FALSE like
is.numeric(), or - variable names through
vars()
- a function that returns TRUE/FALSE like
What do these commands do? Try them out:
# mutate_ifdemo_data %>% mutate_if(is.numeric, as.character) # as.character() is a functiondemo_data %>% mutate_if(is.character, tolower) # tolower() is a functiondemo_data %>% mutate_if(is.double, round, digits=0) # arguments to function can go after# mutate_atdemo_data %>% mutate_at(vars(age:grade), toupper) # toupper() is a functiondemo_data %>% mutate_at(vars(bmi,stweight), log)demo_data %>% mutate_at(vars(contains("race")), str_detect, pattern = "White")# mutate_alldemo_data %>% mutate_all(as.character)Selecting & renaming multiple columns
select_*()&rename_*()are variants ofselect()andrename()- use like
mutate_*()options on previous slide
What do these commands do? Try them out:
demo_data %>% select_if(is.numeric)demo_data %>% rename_all(toupper)demo_data %>% rename_if(is.character, toupper)demo_data %>% rename_at(vars(contains("race")), toupper)The pipe operator %>% revisited
- a function performed on (usually) a data frame or tibble
- the result is a transformed data set as a
tibble - Suppose you want to perform a series of operations on a data.frame or tibble
mydatausing hypothetical functionsf(),g(),h():- Perform
f(mydata) - use the output as an argument to
g():g(f(mydata)) - use the output as an argument to
h():h(g(f(mydata)))
- Perform
One option:
h(g(f(mydata)))A long tedious option:
fout <- f(mydata)gout <- g(fout)h(gout)Using pipes - easier to read:
mydata %>% f() %>% g() %>% h()Why use the pipe?
- makes code more readable
h(f(g(mydata)))can get complicated with multiple arguments- i.e.
h(f(g(mydata, na.rm=T), print=FALSE), type = "mean")
- i.e.
tidyverse way:
demo_data2 <- demo_data %>% na.omit %>% mutate( height_m = sqrt(stweight/bmi), bmi_high = 1*(bmi>30) ) %>% select_if(is.numeric)demo_data2base R way:
demo_data3 <- na.omit(demo_data)demo_data3$height_m <- sqrt(demo_data3$stweight/demo_data3$bmi)demo_data3$bmi_high <- 1*(demo_data3$bmi>30)demo_data3 <- demo_data3[,c("record","bmi","stweight","height_m","bmi_high")]demo_data3Resources - Tidyverse & Data Wrangling
Links
Some of this is drawn from materials in online books/lessons:
- R for Data Science - by Garrett Grolemund & Hadley Wickham
- Modern Dive - An Introduction to Statistical and Data Sciences via R by Chester Ismay & Albert Kim
- A gRadual intRoduction to the tidyverse - Workshop for Cascadia R 2017 by Chester Ismay and Ted Laderas
- "Tidy Data" by Hadley Wickham
Possible Future Workshop Topics?
- reproducible reports in R
- tables
- ggplot2 visualization
- advanced tidyverse: functions, purrr
- statistical modeling in R
Contact info:
Jessica Minnier: minnier@ohsu.edu
Meike Niederhausen: niederha@ohsu.edu
This workshop info:
- Code for these slides on github: jminnier/berd_r_courses
- all the R code in an R script
- answers to practice problems can be found here: html