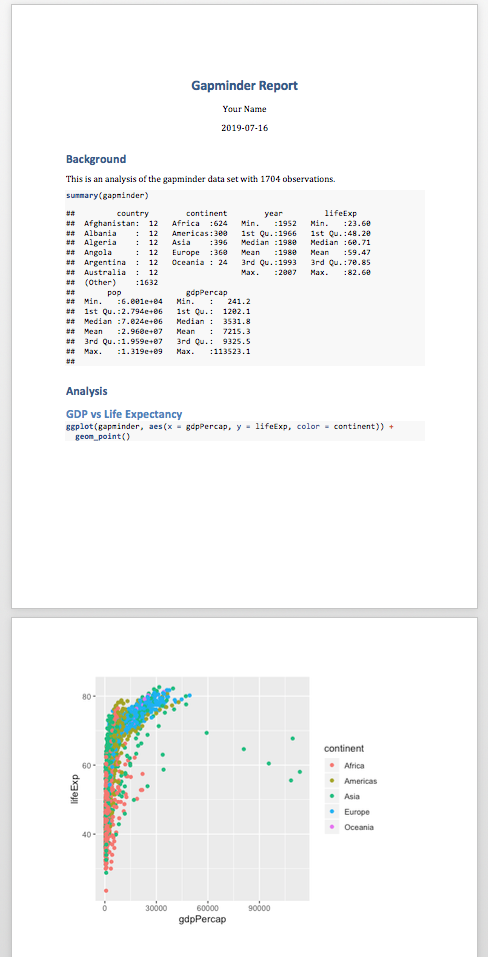
Reproducible Reports with R Markdown
Jessica Minnier, PhD & Meike Niederhausen, PhD
OCTRI Biostatistics, Epidemiology, Research & Design (BERD) Workshop
2019/07/18 & 2019/09/25
slides: bit.ly/berd_rmd
pdf: bit.ly/berd_rmd_pdf
Load files for today's workshop
- Open slides bit.ly/berd_rmd
- Get project folder
- Download zip folder at bit.ly/berd_rmd_zip
- UNZIP completely (right click-> "extract all")
- Open unzipped folder
- Open (double click)
berd_rmarkdown_project.Rproj - Inside RStudio 'Files' tab: click on file
00-install.Rand click "Run" to run all lines of code.
Learning objectives
- Understand how to use literate programming for reproducible research
- Basics of Markdown language
- Learn how to create R Markdown files with code and markdown text
- Turn R Markdown files into html, pdf, Word, or presentation files
- Learn about reproducible project workflows
- (If time allows) Learn some additional R Markdown tips
Why Reproducibility?
- Evidence your results are correct.
- Allow others to use our methods and results.
"An article about computational results is advertising, not scholarship. The actual scholarship is the full software environment, code and data, that produced the result."
-- (Claerbout and Karrenbach 1992)
Your closest collaborator is you six months ago, but you don't reply to emails.
Types of Reproducibility
Computational reproducibility: detailed information is provided about
- code, software, hardware and implementation details.
Empirical reproducibility: detailed information is provided about
- non-computational empirical scientific experiments and observations [data].
Statistical reproducibility: detailed information is provided about
- the choice of statistical tests, model parameters, threshold values, etc.
R Opensci Reproducibility Guide
Software tool for reproducibility: Literate Programming
"These tools enable writing and publishing self-contained documents that include narrative and code used to generate both text and graphical results.
In the R ecosystem, knitr [R markdown] and its ancestor Sweave used with RStudio are the main tools for literate computing. Markdown or LaTeX are used for writing the narrative, with chunks of R code sprinkled throughout the narrative. IPython is a popular related system for the Python language, providing an interactive notebook for browser-based literate computing."
R Opensci Reproducibility Guide
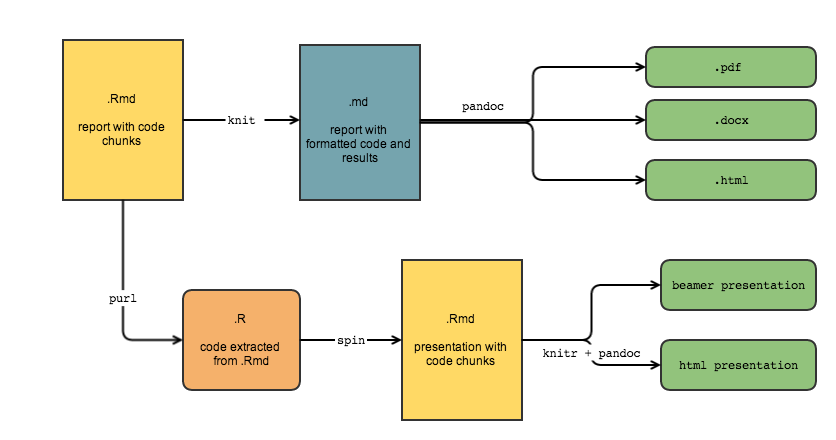
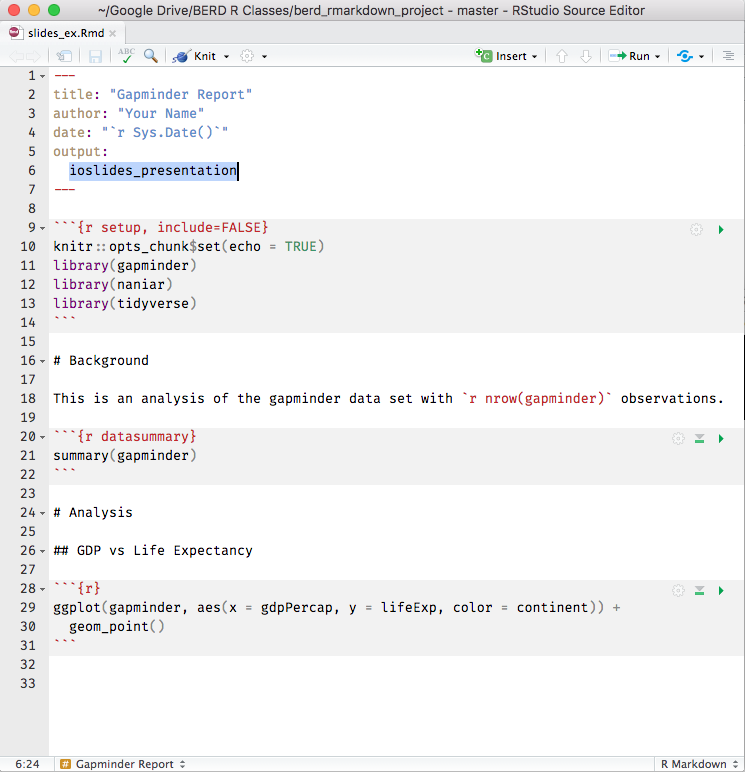
R Markdown = .Rmd file = Code + text
knitr is a package that converts .Rmd files containing code + markdown syntax to a plain text .md markdown file, and then to other formats (html, pdf, Word, etc)
 ⇒
⇒
knitr converts .Rmd -> .md (behind the scenes)

knitr converts .Rmd -> .md -> .html
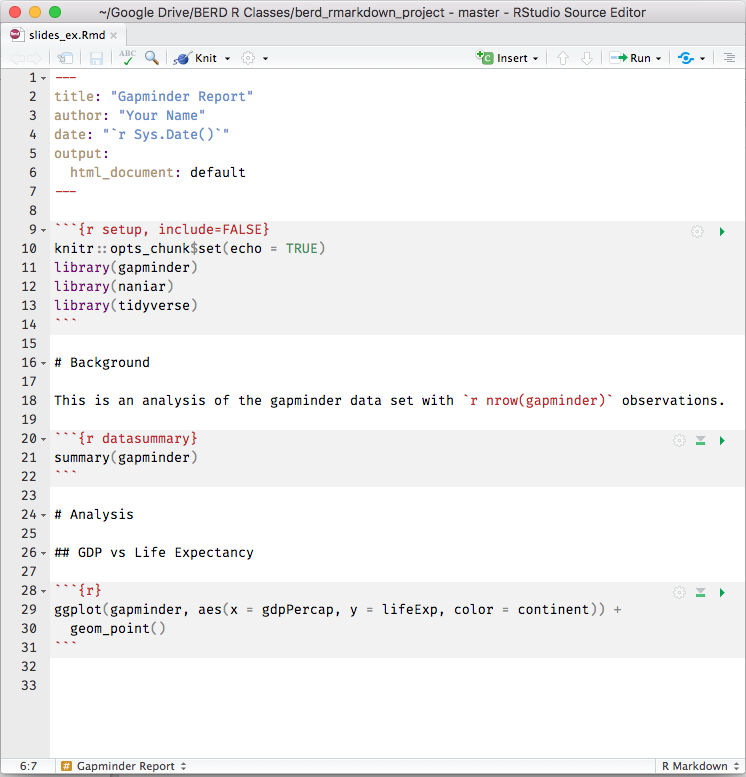

knitr converts .Rmd -> .md -> .pdf
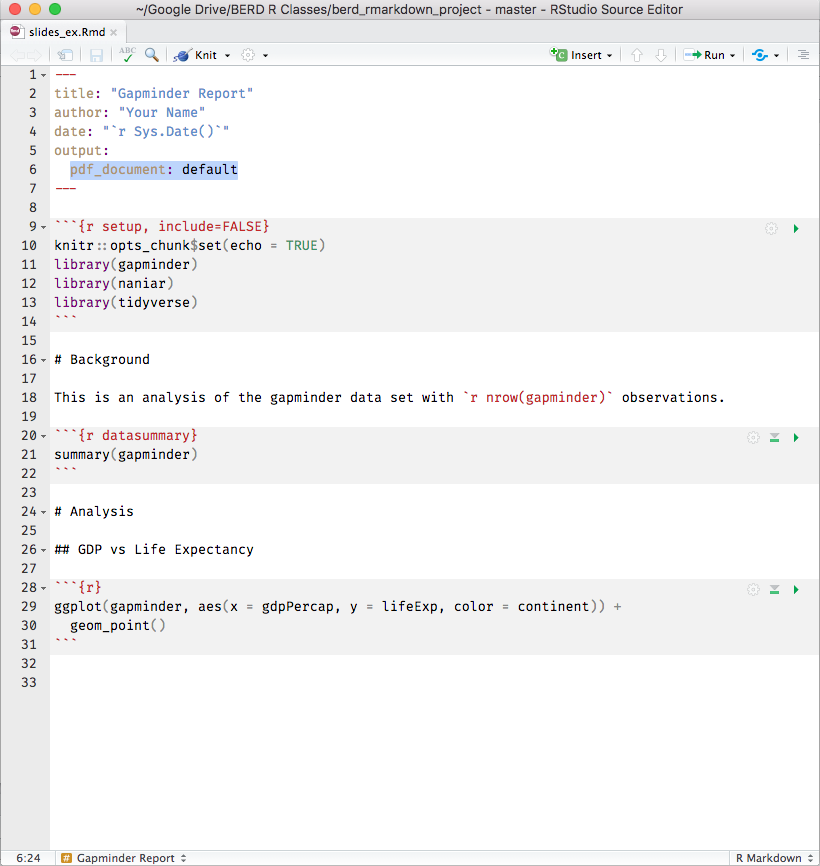
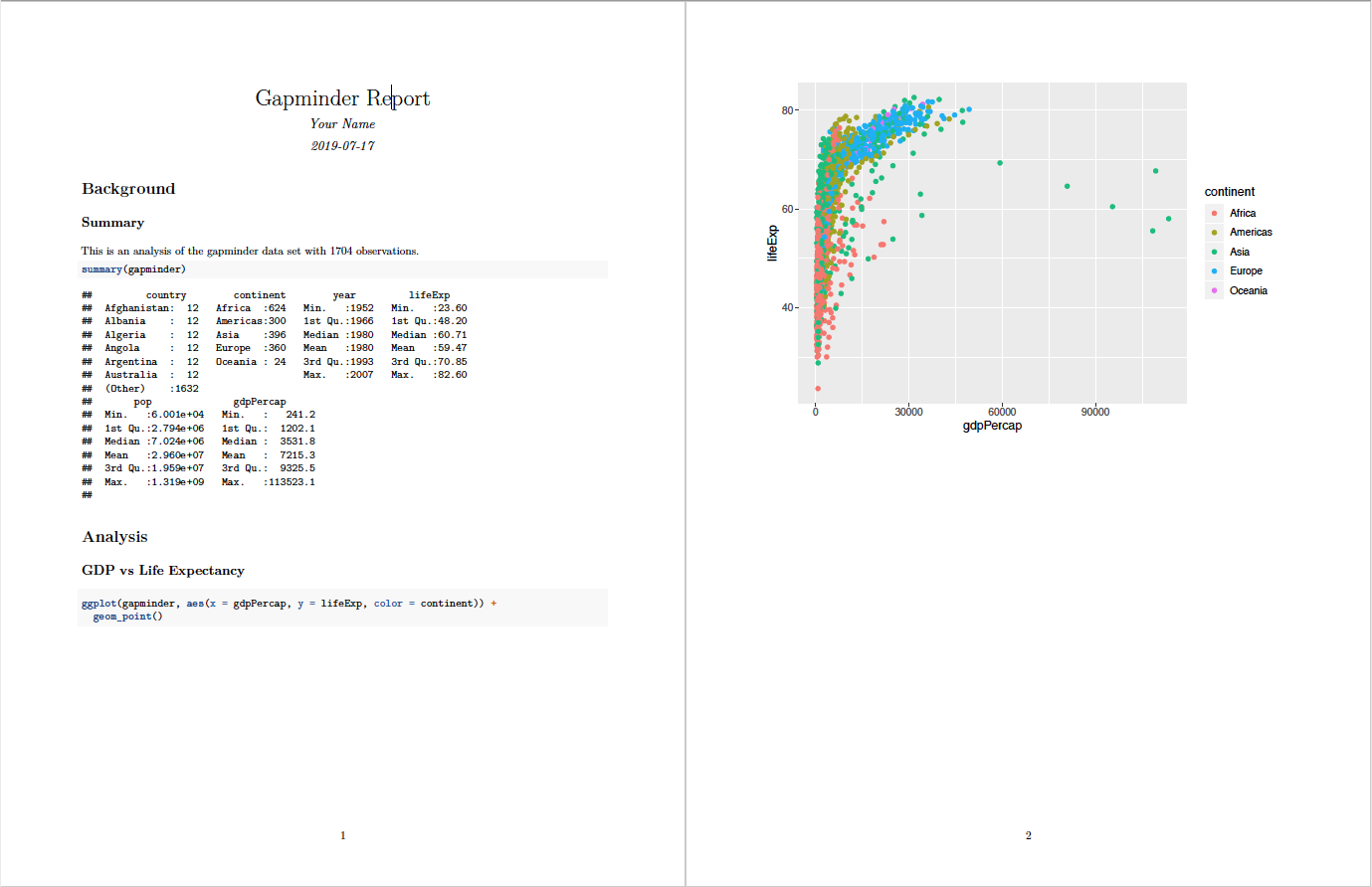
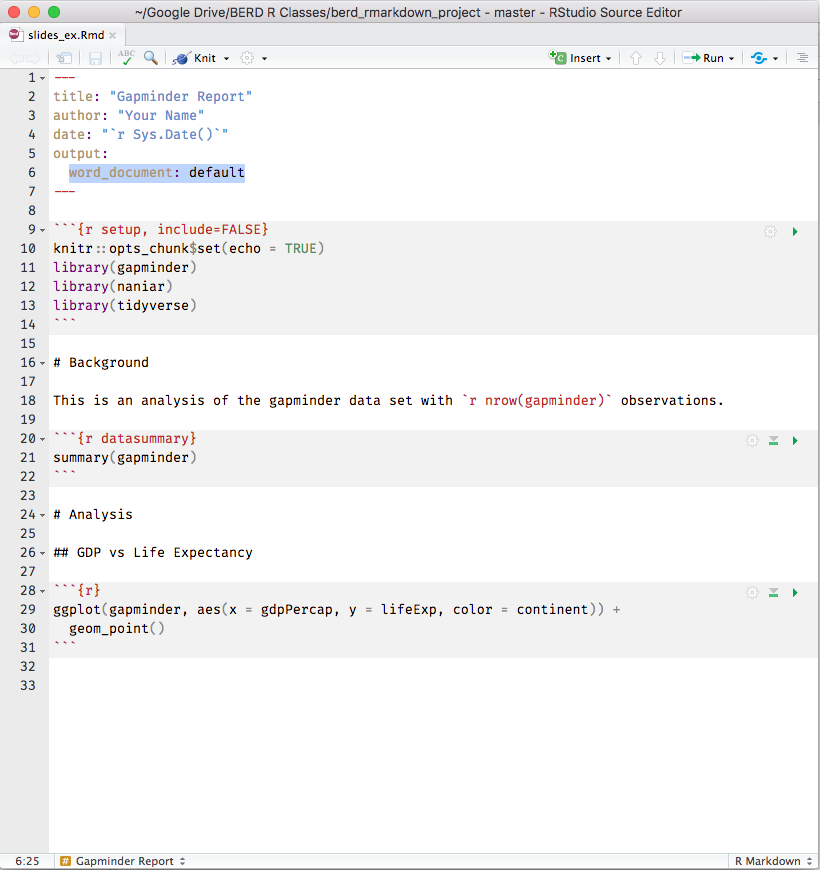
knitr converts .Rmd -> .md -> .doc
knitr converts .Rmd -> .md -> slides
Good practices in RStudio
Use projects (read this)
- Create an RStudio project for each data analysis project
- A project is associated with a directory folder
- Sets working directory
- Keep data files there
- Keep scripts there; edit them, run them in bits or as a whole
- Save your outputs (plots and cleaned data) there
- Only use relative paths, never absolute paths
- relative (good):
read_csv("data/mydata.csv") - absolute (bad):
read_csv("/home/yourname/Documents/stuff/mydata.csv")
- relative (good):
Advantages of using projects
- standardize file paths
- keep everything together
- a whole folder can be shared and run on another computer
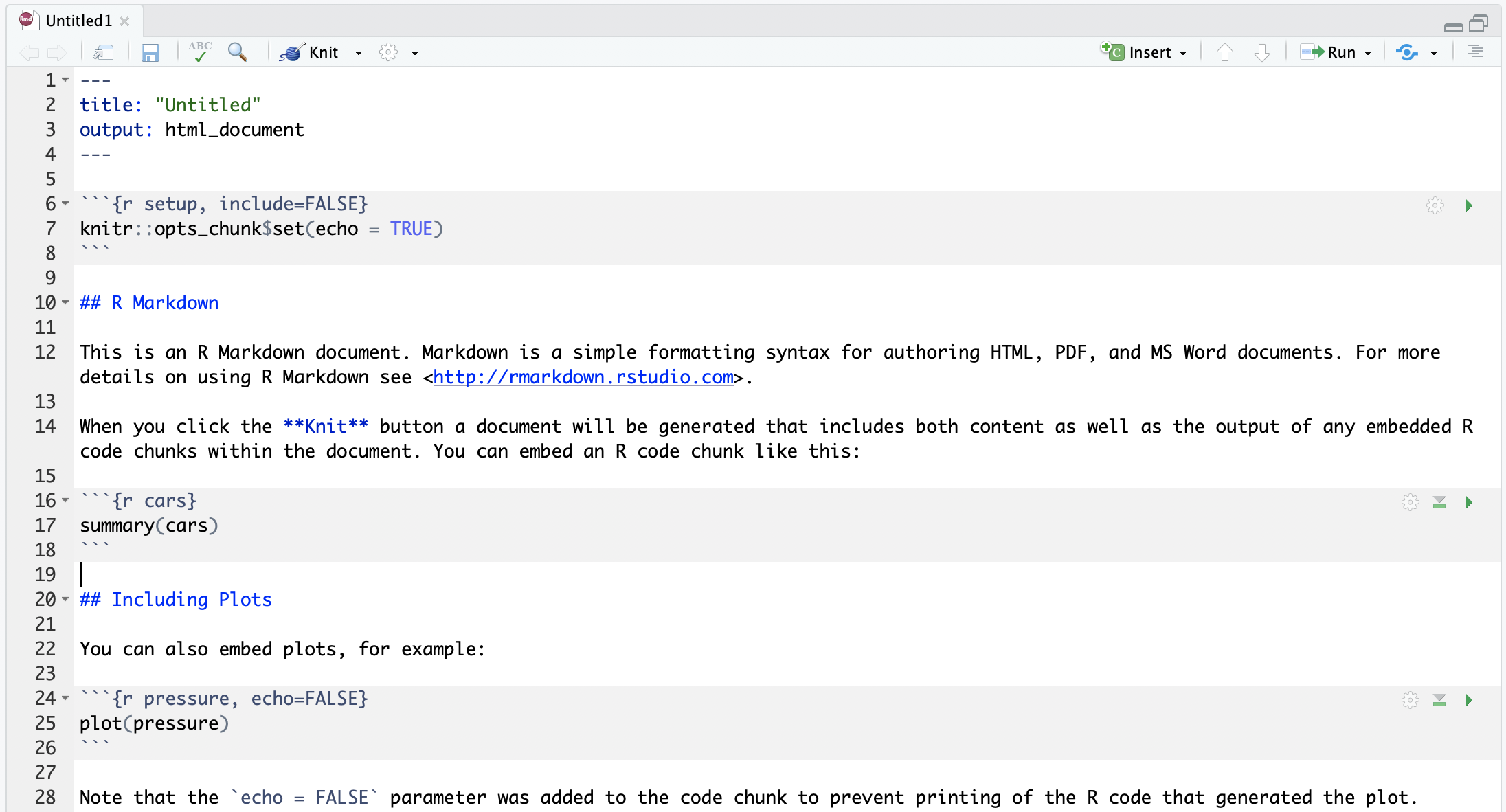
Create an R Markdown file (.Rmd)
Two options:
- click on File → New File → R Markdown... , or
- in upper left corner of RStudio click on
 →
→ 
You should see the following text in your editor window:
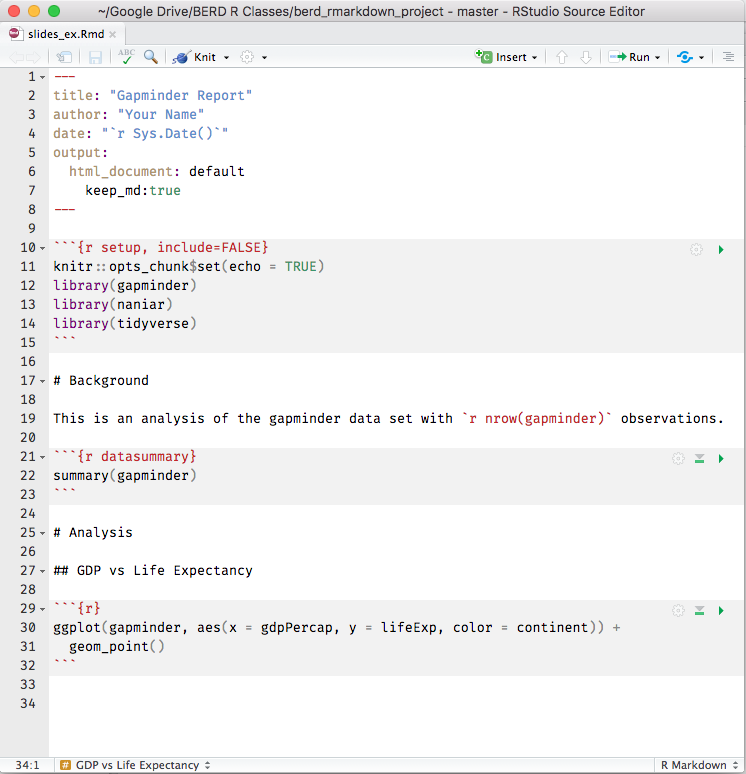
Knit the .Rmd file
Before knitting the .Rmd file, you must first save it.
To knit the .Rmd file, either
- click on the knit icon
 at the top of the editor window
at the top of the editor window - or use keyboard shortcuts
- Mac: Command+Shift+K
- PC: Ctrl+Shift+K
- or use the
render()command in Console - See Extensions section for details
A new window will open with the html output.
Remark:
- The template .Rmd file that RStudio creates will knit to an html file by default
- Later we will go over knitting to other file types
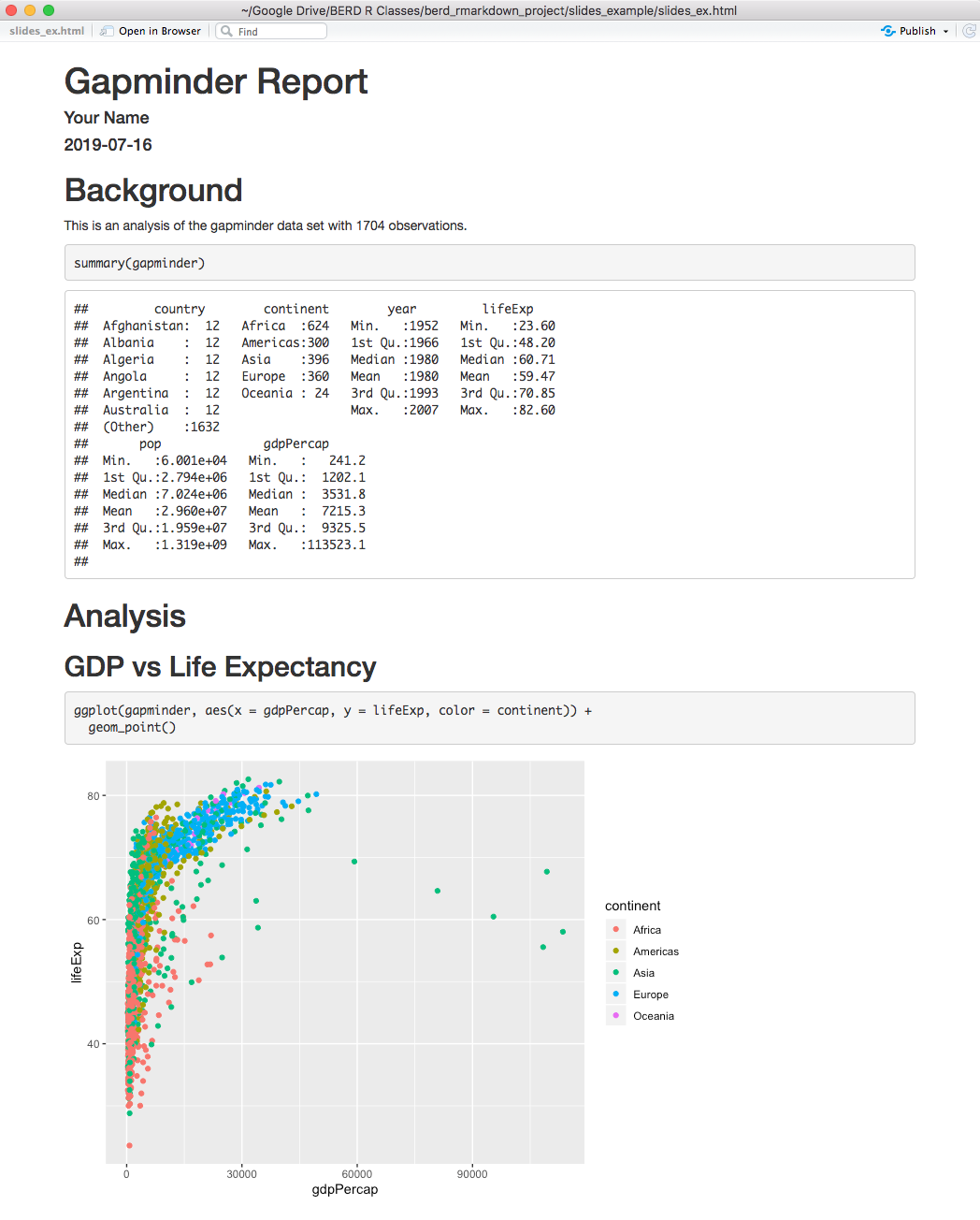
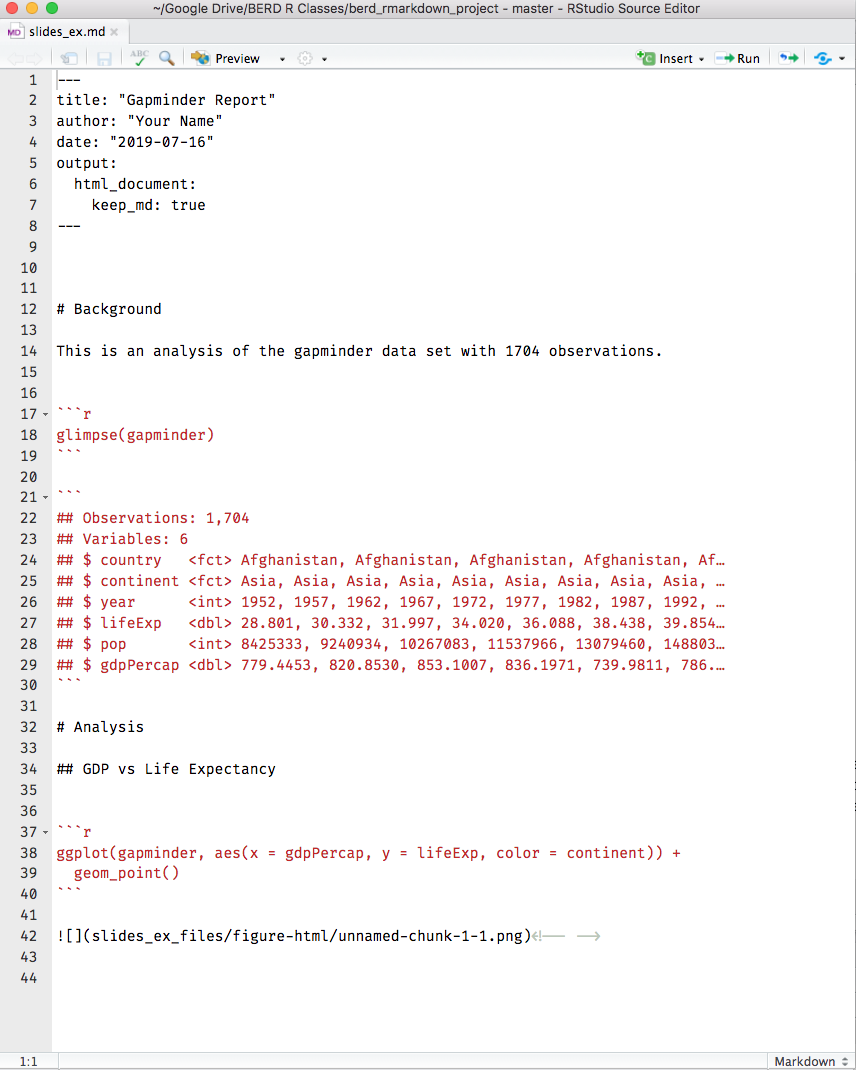
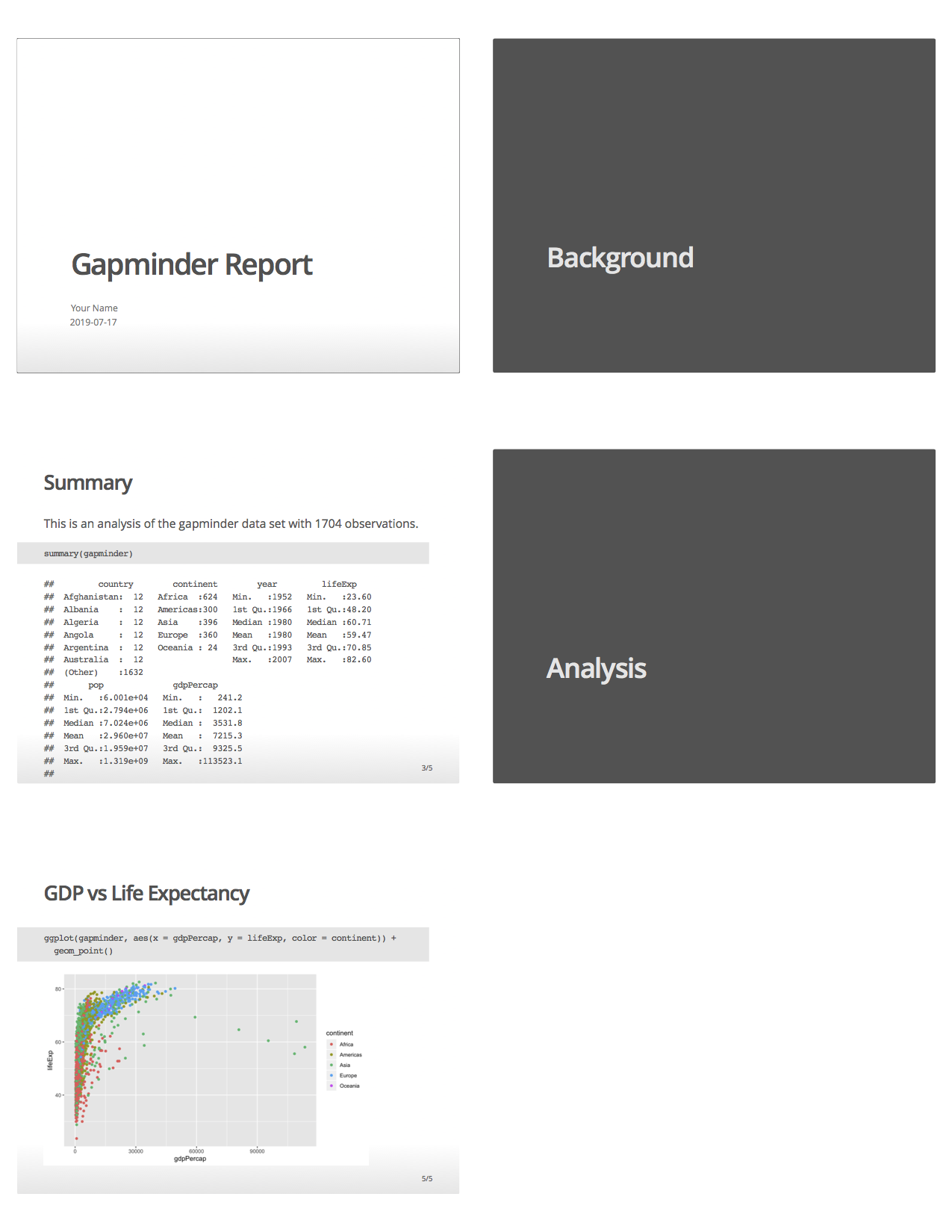
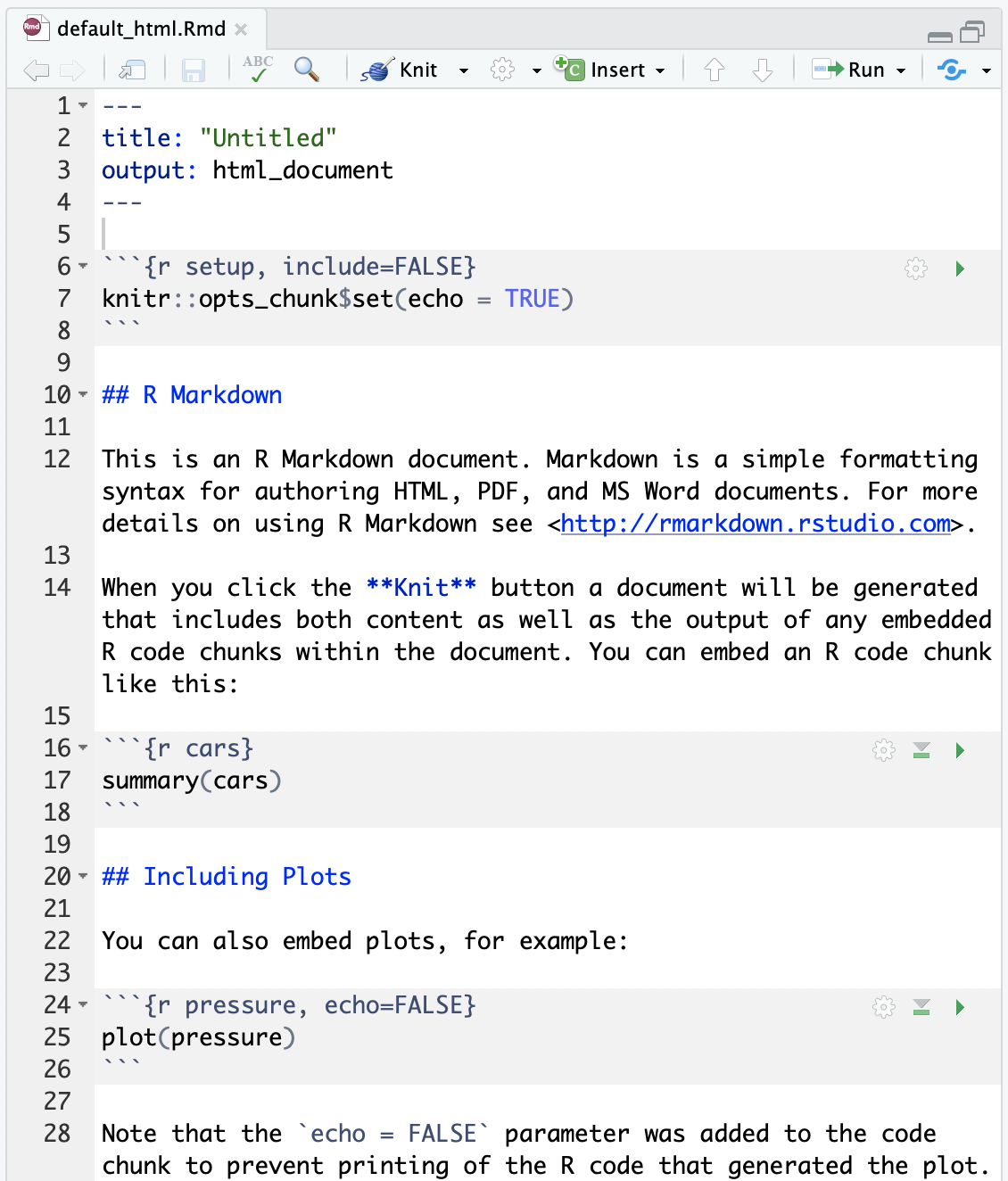
Compare the .Rmd file with its html output
.Rmd file
html output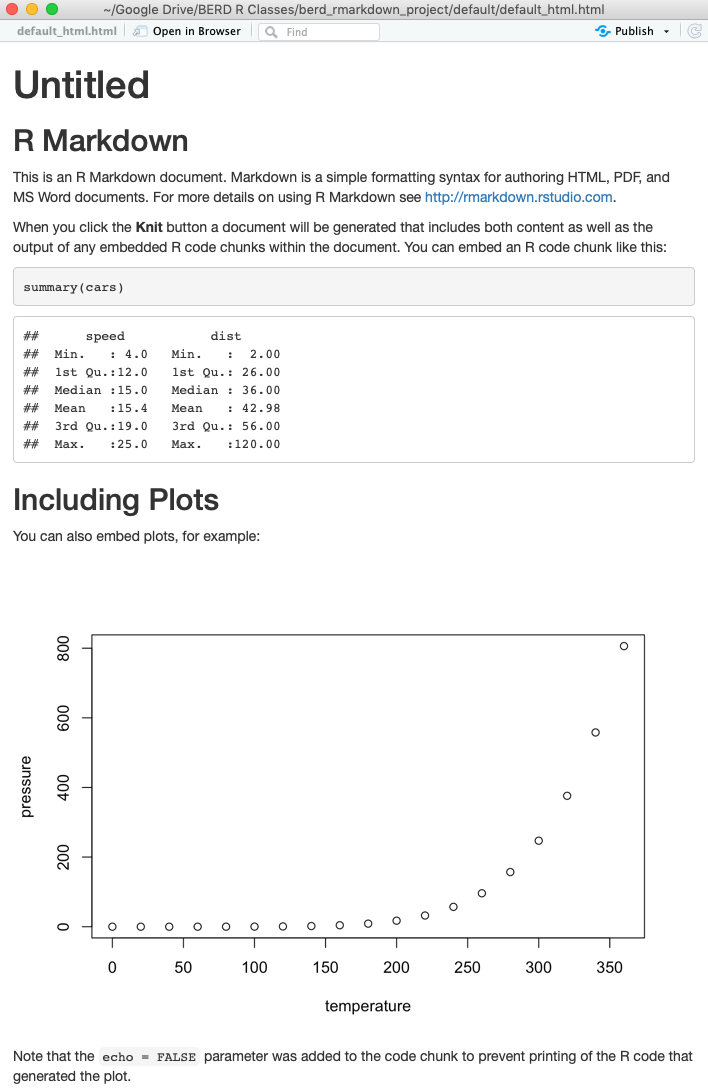
Compare the .Rmd file with its html output
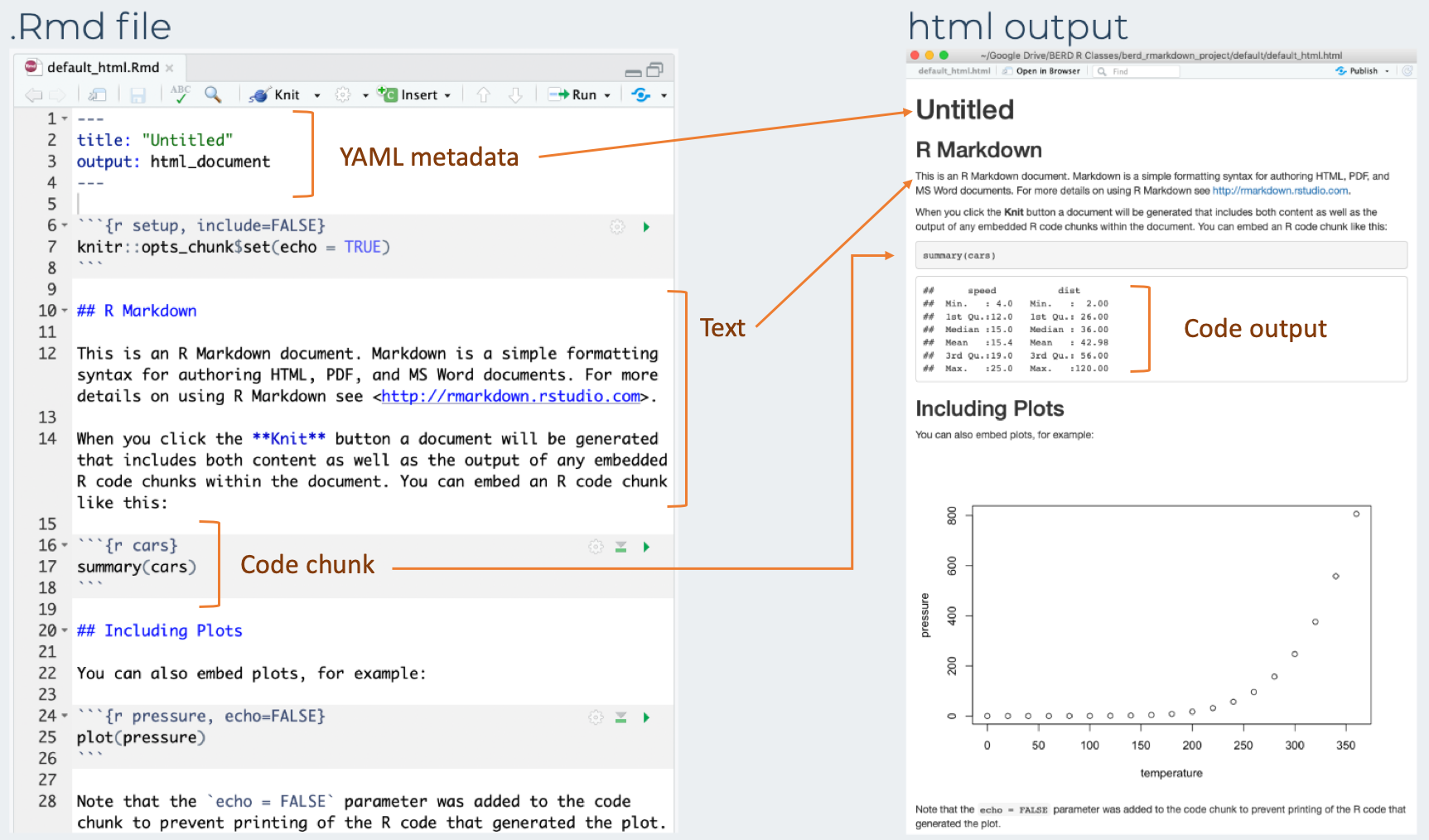
3 types of R Markdown content
- Text
- Code chunks
- YAML metadata
Formatting text
- Markdown is a markup language similar to html or LaTeX
- All text formatting is specified via code
Text in editor:

Output:

Headers
- Organize your documents using headers to create sections and subsections
- Later in the workshop we will cover
- automatically numbering headers in output file for easy reference
- easily creating a TOC based on the header names
Text in editor:

Output:

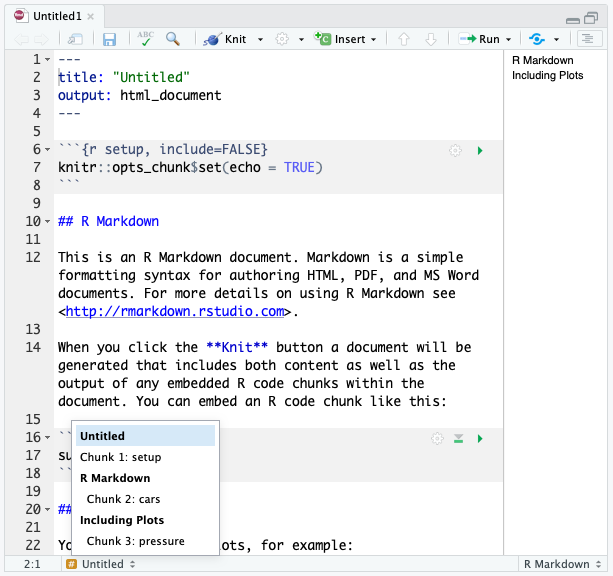
RStudio tip
You can easily navigate through your .Rmd file if you use headers to outline your text
Unnumbered lists
Text in editor:

- This is an unnumbered list
- with sub-items
- and sub-sub-items,
- or even deeper.
- and sub-sub-items,
- with sub-items
- You can use characters *, +, and - to create lists.
- The order of the
- characters is not important
- and characters can be repeated.
- characters is not important
- The order of the
What is important is the spacing!
- indent each
- sub-level with a tab and make sure
- there is a space between the character starting the list and the first bit of text, *otherwise the text won't be a new bullet in the list
Numbered lists
Text in editor:

Output:

Math, horizontal rule, and hyperlinks
Text in editor:
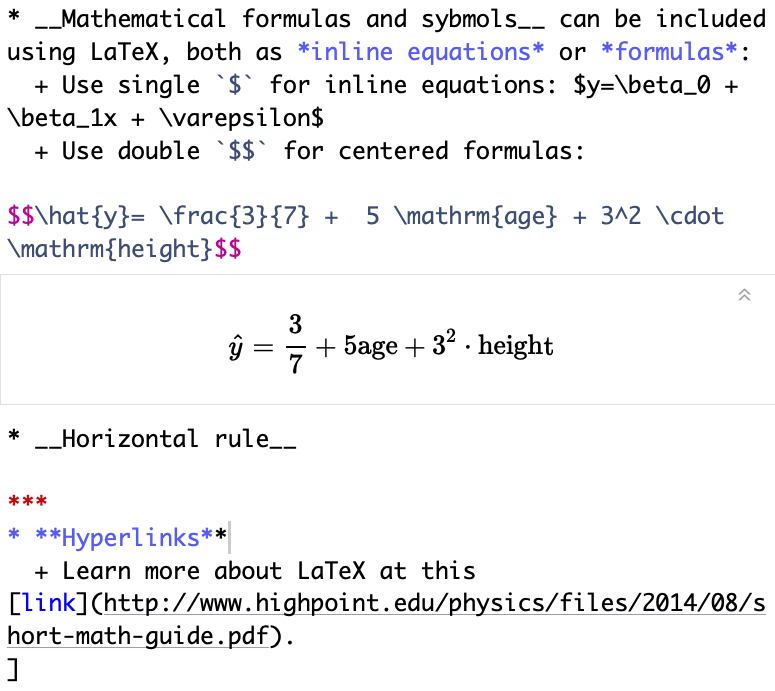
Output:
- Mathematical formulas and symbols can be included using LaTeX, both as inline equations or formulas:
- Use single
$for inline equations: y=β0+β1x+ε - Use double
$$for centered formulas:
- Use single
^y=37+5age+32⋅height
- Horizontal rule
- Hyperlinks
- Learn more about LaTeX at this link.
Insert images
Text in editor:
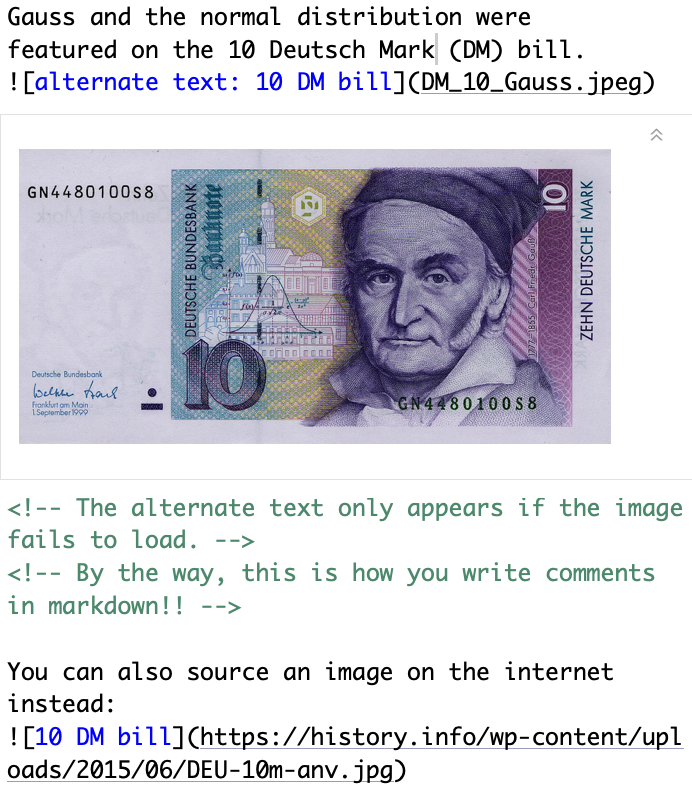
Output:
Gauss and the normal distribution were
featured on the 10 Deutsch Mark (DM) bill.

You can also source an image on the internet instead:

Tables created manually
Later we will use R code to create tables from data.
We can create tables using Markdown as well:
Text in editor:
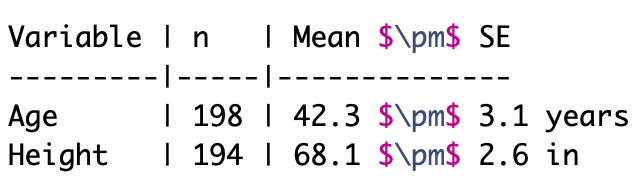
Output:
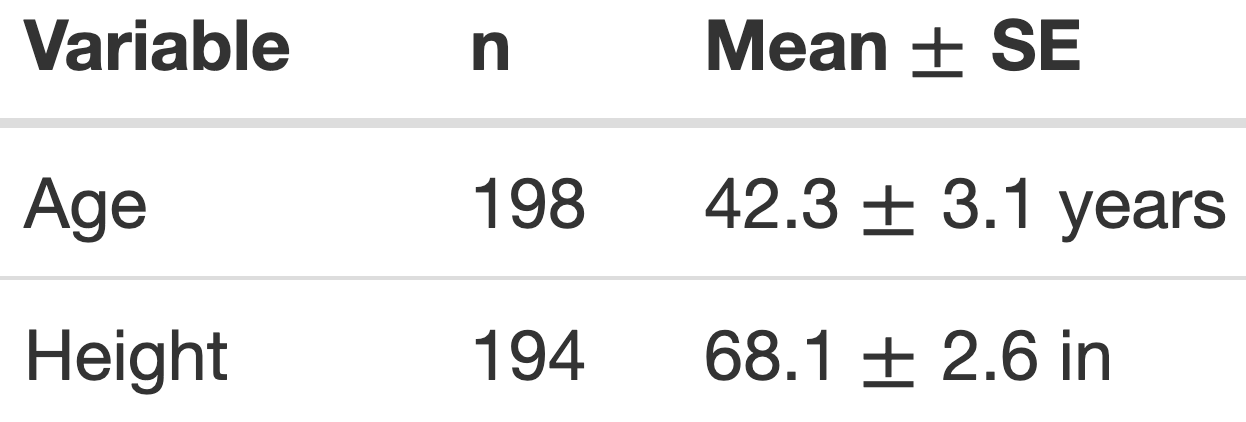
- We do not recommend creating tables where the numbers are hard-coded
- since they are not reproducible!
Spell check
Alas, there are no autmatik sepll chekc to katch you're tipos and grammR.
- You can manually do a spell check by clicking on the
 icon above the editor window.
icon above the editor window. - There is no built-in grammar check in RStudio.
- The
gramrpackage is an available RStudio Addin.
- The
Practice!
Create an .Rmd file with file name example1.Rmd that creates the html output to the right.
- Hint: The first line is not a header.

3 types of R Markdown content
- Text
- Code chunks
- YAML metadata
Data description: Fisher's (or Anderson's) Iris data set
- n = 150
- 3 species of Iris flowers (Setosa, Virginica, and Versicolour)
- 50 measurements of each type of Iris
- variables:
- sepal length, sepal width, petal length, petal width, and species
Can the flower species be determined by these variables?


Code chunks
Chunks of R code start with ```{r} and end with ``` .
For example, the chunk  produces the output
produces the output
summary(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300 Median :5.800 Median :3.000 Median :4.350 Median :1.300 Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800 Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500 Species setosa :50 versicolor:50 virginica :50Create a code chunk
Code chunks can be created by either
Clicking on
 →
→  at top right of editor window, or
at top right of editor window, orKeyboard shortcut
- Mac: Command + Option + I
- PC: Ctrl + Alt + I
Chunk options- most common
Text in editor:

No options specified: see both code and output
mean(iris$Sepal.Length)[1] 5.843333echo determines whether the R code is displayed or not. The default is TRUE. When set to FALSE, the code is not displayed in the output:
[1] 5.843333eval determines whether the R code is run or not. The default is TRUE. When set to FALSE, the code is not run but is displayed in the output:
mean(iris$Sepal.Length)More chunk options
Text in editor:

Output:
include determines whether to include the R chunk in the output or not. The default is TRUE. When set to FALSE, the chunk is run but we do not see the code or its output (note that nothing is displayed below):
Setting
include=FALSEis useful when you have R code that you want to run, but do not want to display either the code or its output.See the R Markdown cheatsheet for more chunk options.
Inline code
- You can also report R code output inline with the text
- R code is not shown in this case
Text in editor:
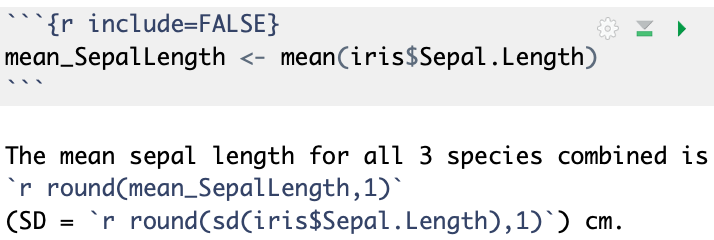
Output:
The mean sepal length for all 3 species combined is 5.8 (SD = 0.8) cm.
- The code above is an example of where
include=FALSEis used a chunk option to evaluate the code but not show the code or its output.- It saves the mean as
mean_SepalLength, which can then be used later on.
- It saves the mean as
- For the standard deviation, the inline code did the calculation.
- Thus it was not necessary to first save the mean as a variable.
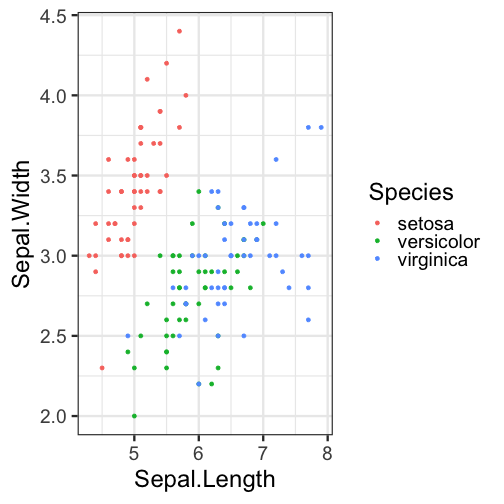
Figures
Text in editor:
- Figure dimensions specified with
fig.widthandfig.height - Figure name specified by the chunk label
- The figure created by the chunk above is called
Sepal_WidthVsHeight-1.png - Chunk names must be unique!
- The figure created by the chunk above is called
echo=FALSEwas used to hide the code and only display the figure
Output:
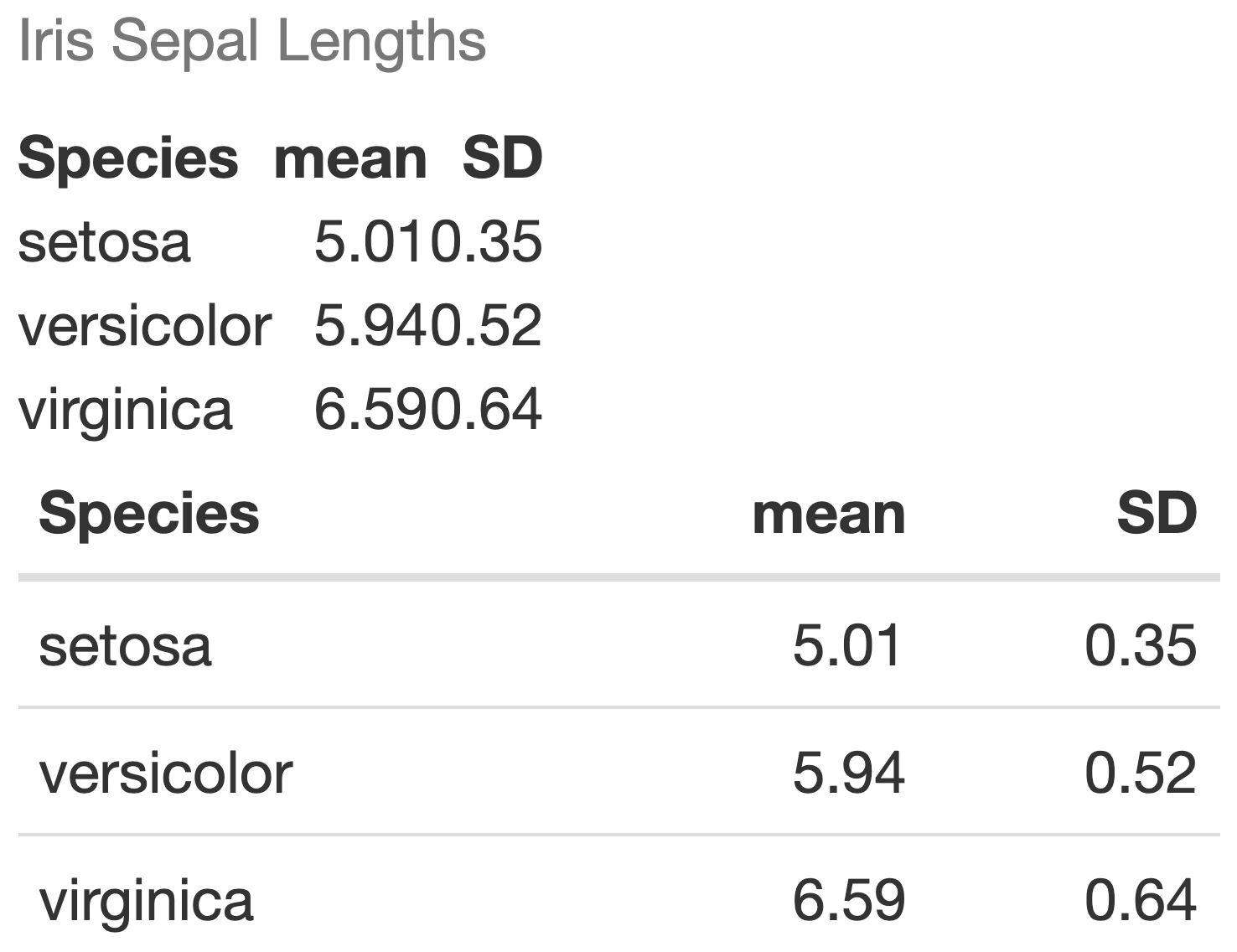
Tables - with no formatting
- Below we create a summary table with the mean and SD of sepal lengths
- The table is displayed with no special formatting
table_sepal_length <- iris %>% group_by(Species) %>% summarize(mean = mean(Sepal.Length), SD = sd(Sepal.Length))table_sepal_length# A tibble: 3 x 3 Species mean SD <fct> <dbl> <dbl>1 setosa 5.01 0.3522 versicolor 5.94 0.5163 virginica 6.59 0.636Tables - with kable
- The
kablecommand from theknitrpackage has some basic formatting options- html tables: harder to read due to squished spacing; can include caption
- markdown tables: nicer formatting; width = page width
Text in editor:

Output:
Tables - use kableExtra for more formatting options
Text in editor:

Output:
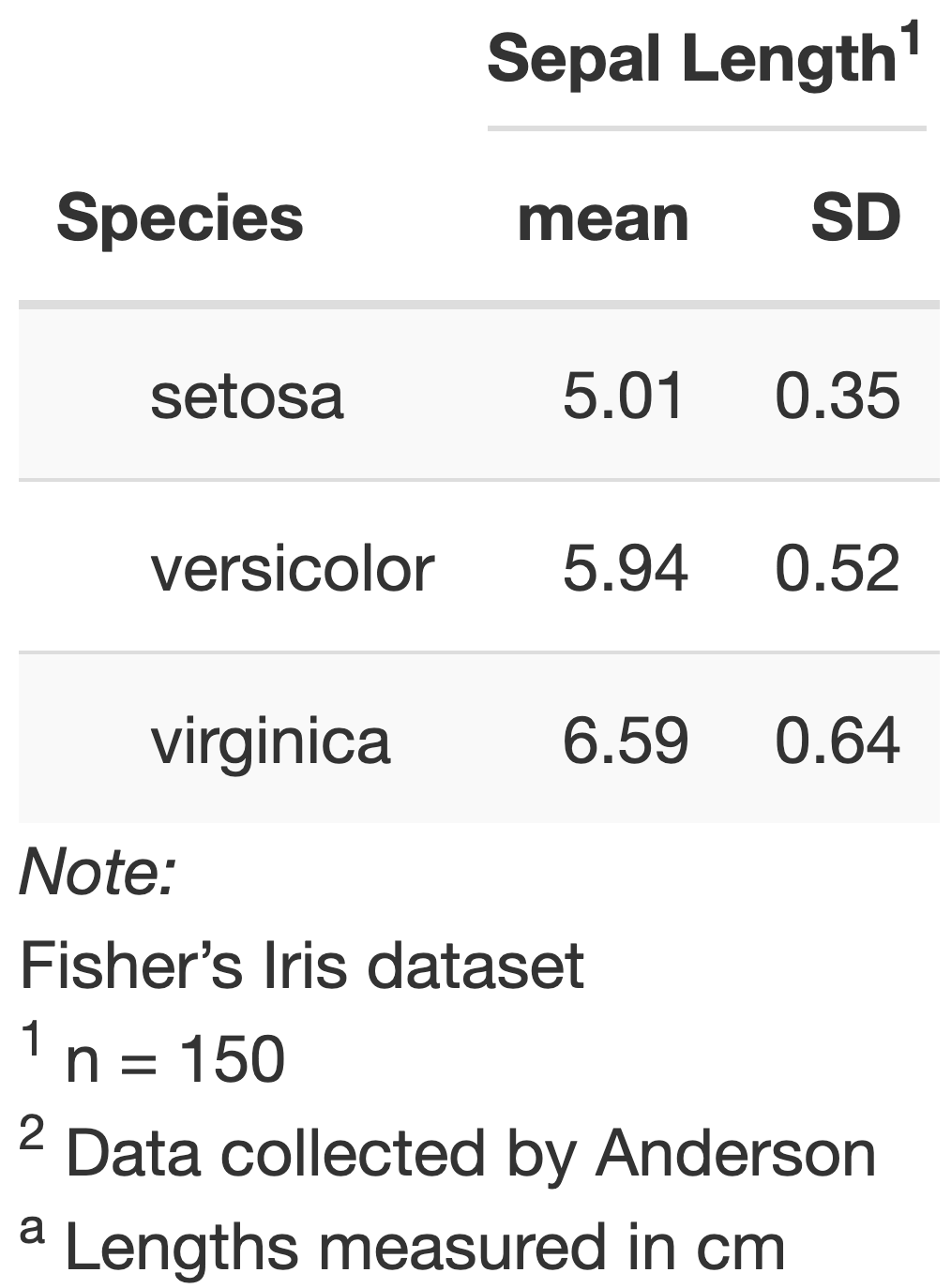
See Hao Zhu's webpage for many, many more kableExtra options.
Global chunk options
- You can set global chunk options that are applied to all chunks in the .Rmd file
- Set global options in a chunk at the beginning of the .Rmd file
- The template .Rmd file already includes a chunk labeled
setup - Add more options as desired to this chunk
- Options are added within the
knitr::opts_chunk$set(...)command - Any of the many chunk options can be set in the
setupchunk

fig.pathsets the folder name where figures generated by the .Rmd file will be saved- See the R Markdown cheatsheet for more chunk options.
Practice! (part 1)
Edit the file example2/example2.Rmd to create html output that matches example2/example2_output.html shown below.
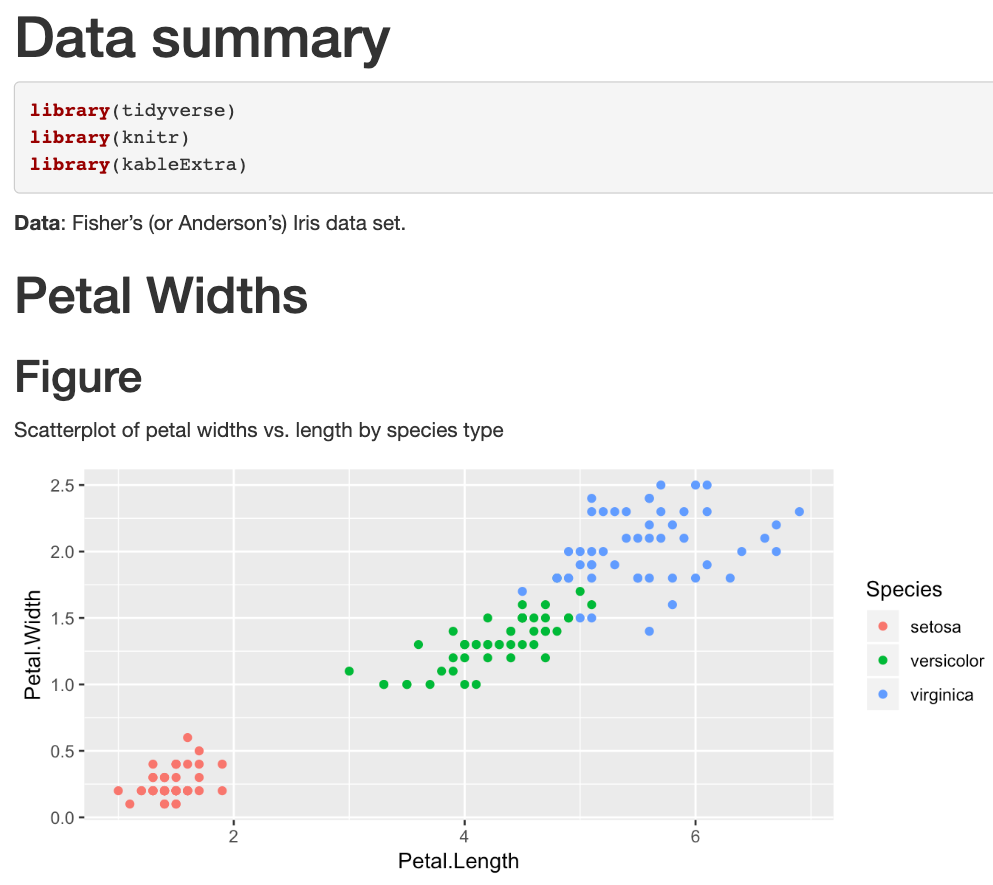
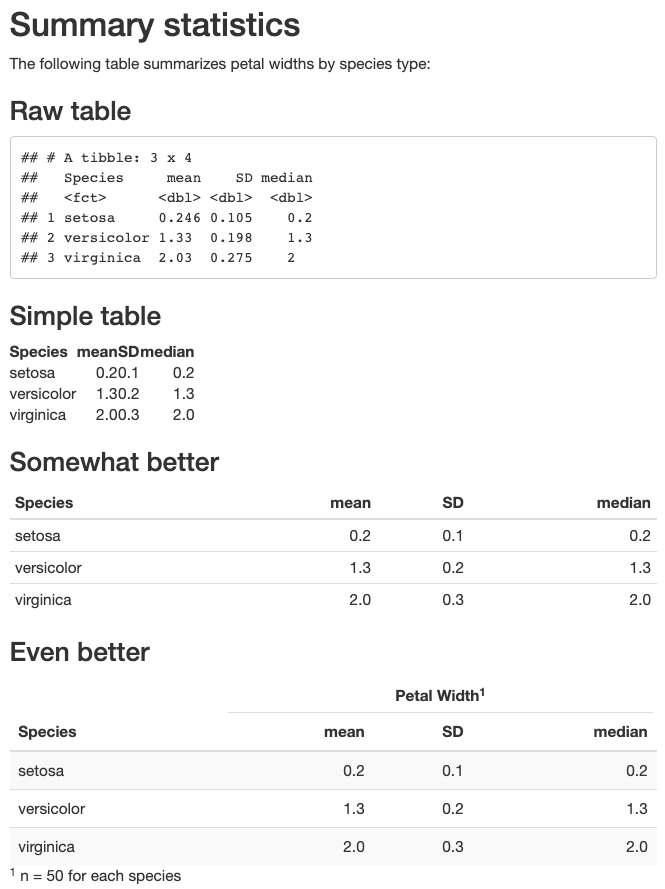
Practice! (part 2)
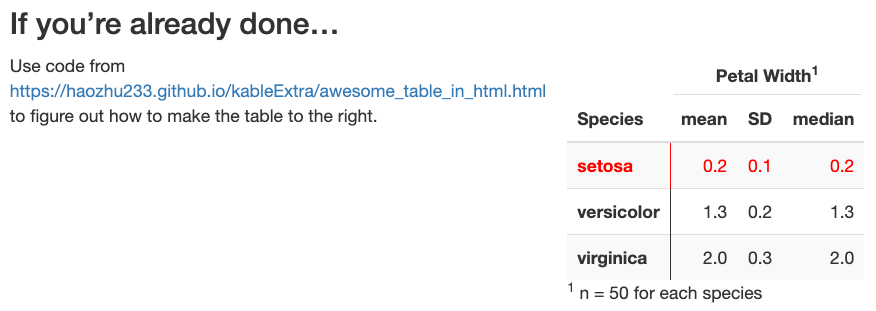
Create the table output shown below and at the end of example2/example2_output.html (code link)
3 types of R Markdown content
- Text
- Code chunks
- YAML metadata
YAML metadata
Many output options can be set in the YAML metadata, which is the first set of code in the file starting and ending with ---.
- YAML is an acronym for YAML Ain't Markup Language
- It sets the configuration specifications for the output file
- For more details about YAML in general, see the YAML Wikipedia page
Set the title, author, and date that appear at the top of the output file
Text in editor:
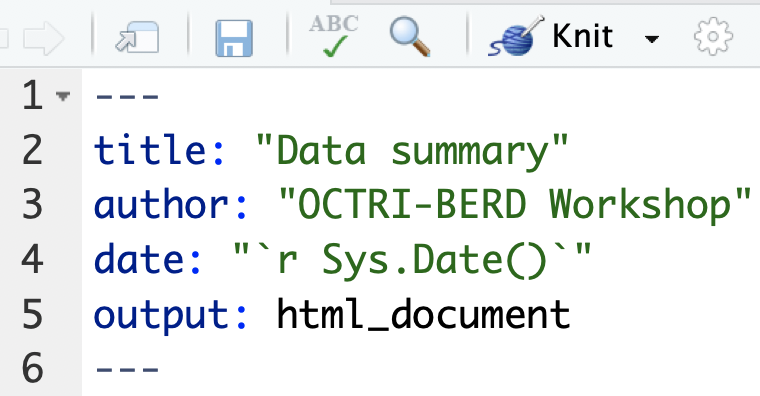
Output:

Numbered sections & clickable table of contents
Text in editor: (example3a.Rmd)
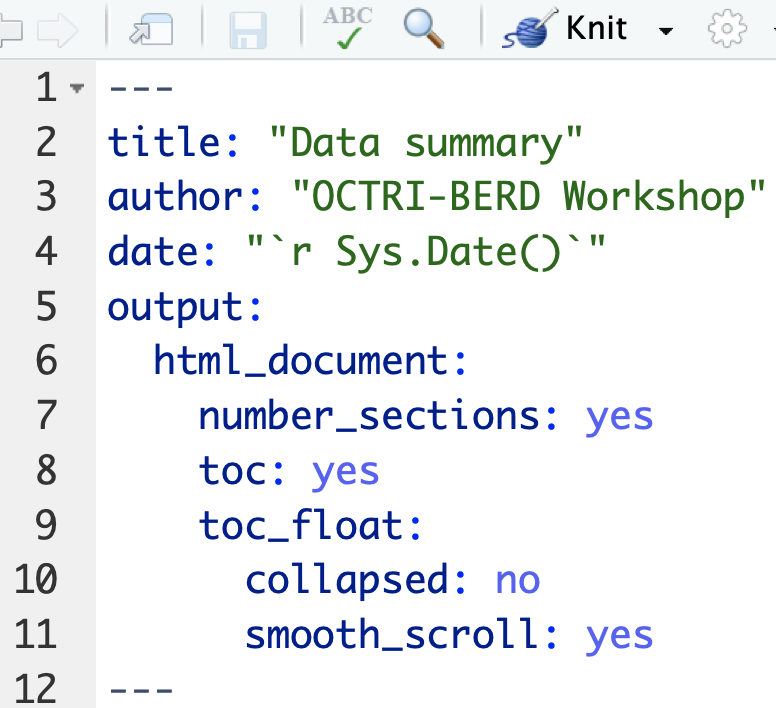 Try out
Try out collapsed: yes and smooth_scroll: no
Output: (example3a.html)
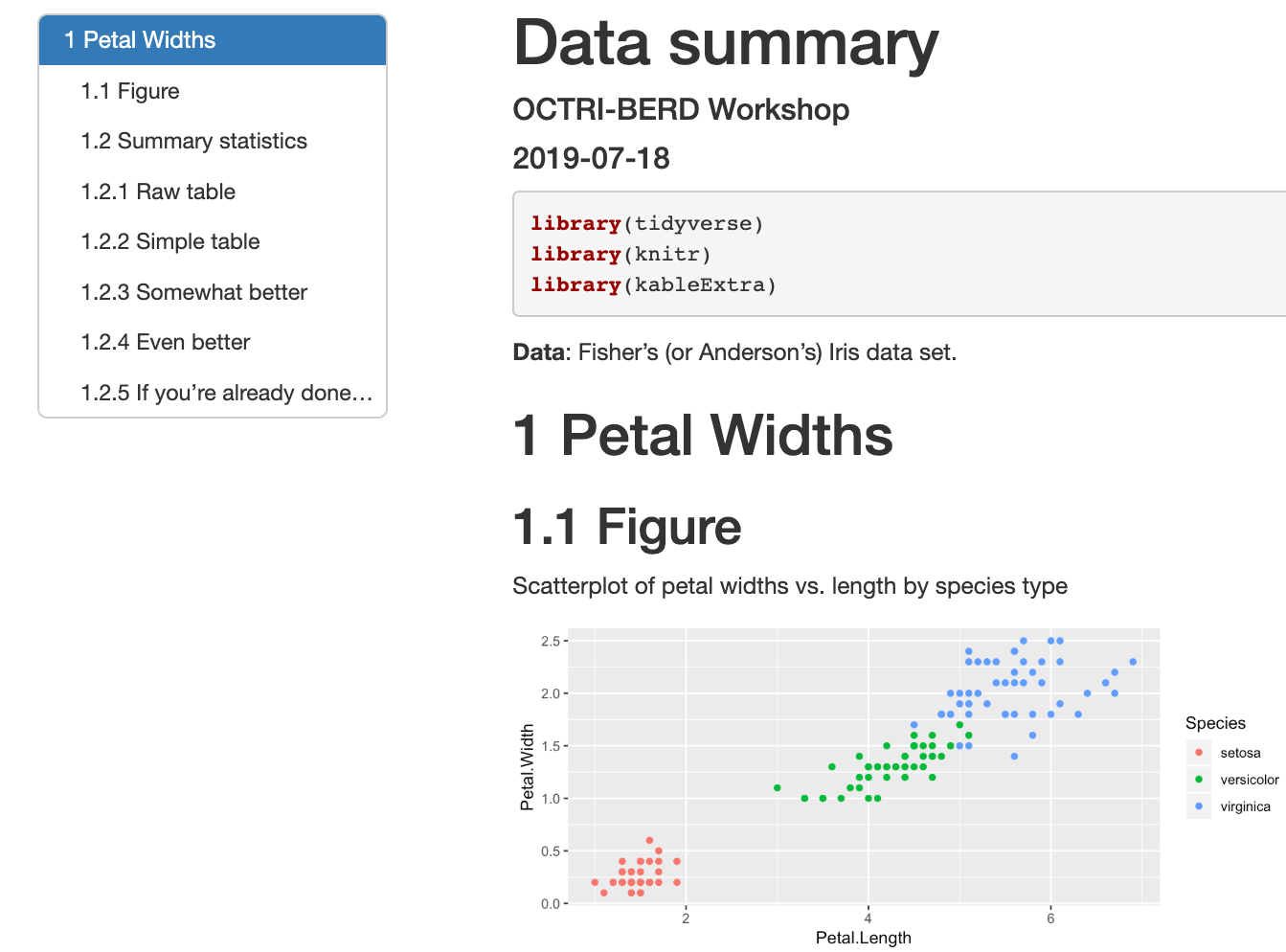
Themes
- There are 12 themes to choose from without installing additional packages
- See http://www.datadreaming.org/post/r-markdown-theme-gallery/ for examples
Text in editor: (example3b.Rmd)
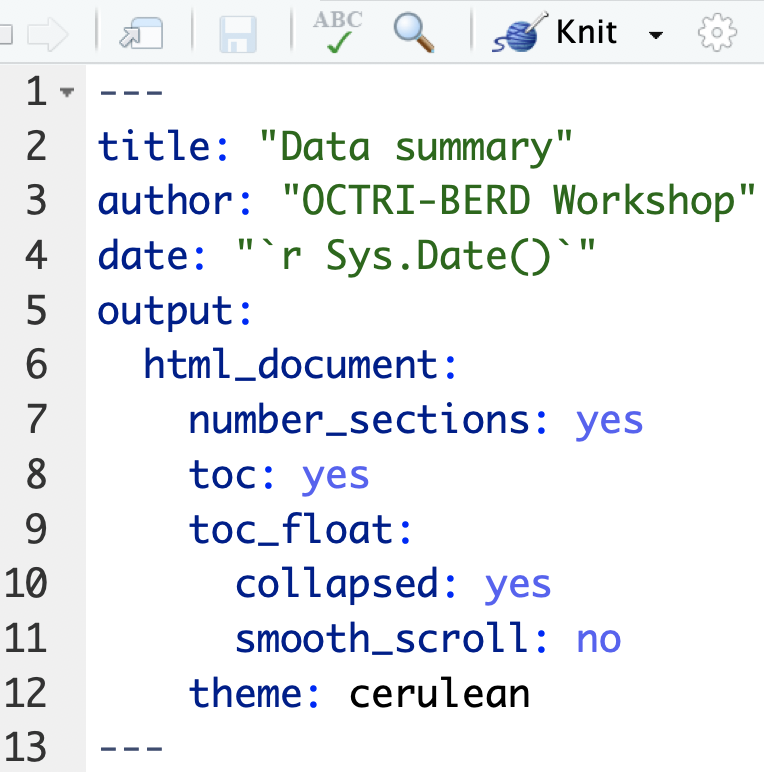
Output: (example3b.html)
Code folding
- Code folding creates buttons in the output html file that lets users choose whether they want to see the R code or not
- This only applies to R code from chunks with
echo = TRUE
- This only applies to R code from chunks with
code_folding: hideall R code hidden by default; user must click Code button to see Rcode_folding: showall R code shown by default; user must click Code button to hide R- See https://bookdown.org/yihui/rmarkdown/html-document.html#code-folding for more info

Word documents
- Not many YAML options
- Cannot include html code or html-specific commands
Text in editor: (Word_example3.Rmd)
Output: (Word_example3.docx)
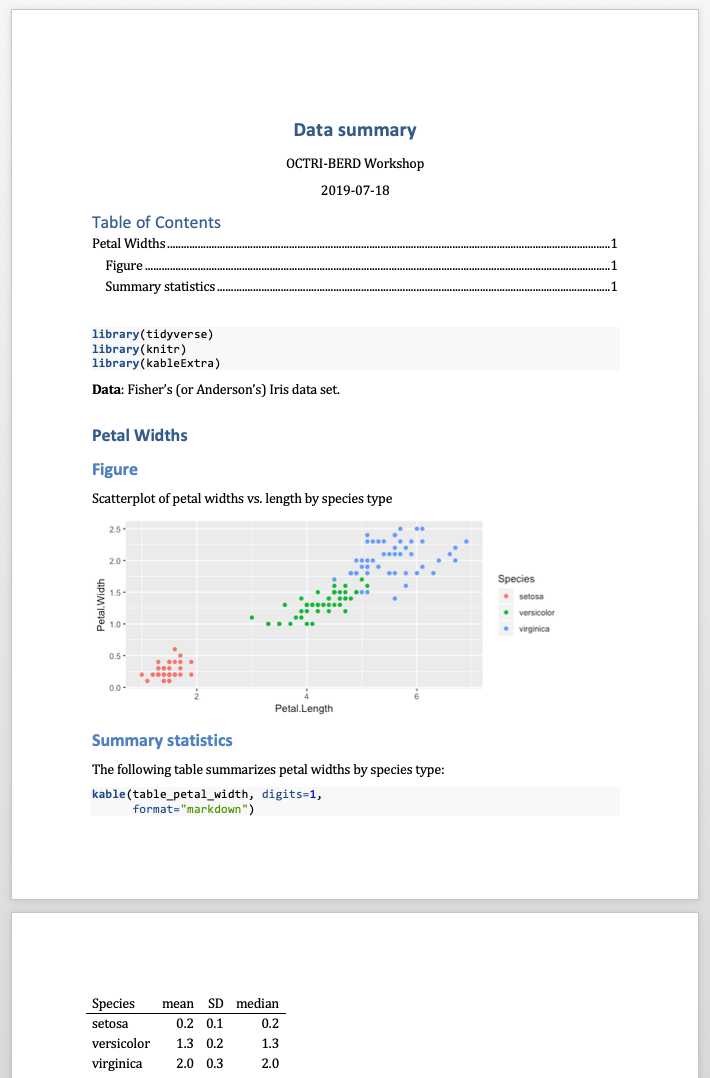
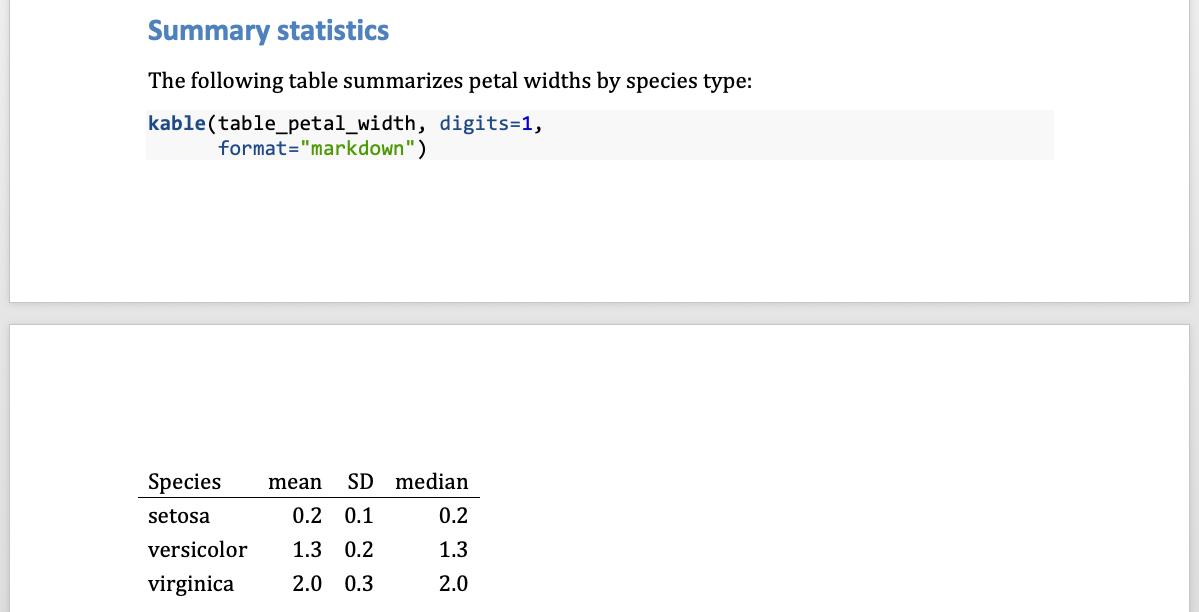
Word documents - tables options limited
- Cannot use
kableExtrapackage options kablecan be used
Word documents - using a style file
- Create a Word doc with preferred formatting
- font types and sizes, margins, header colors, etc.
YAML with code to include style file:
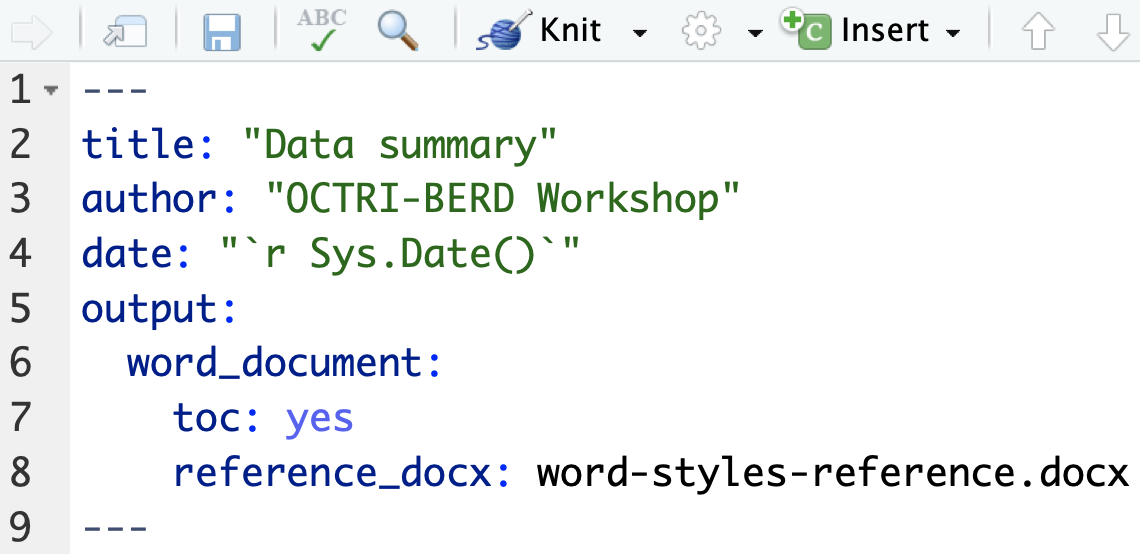
Sample style file: (word-styles-reference.docx)
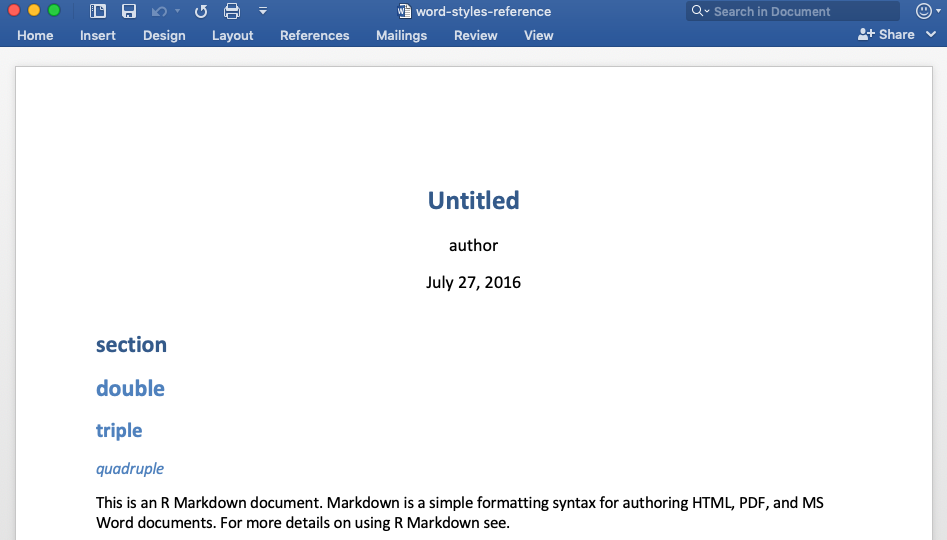
The Word doc created by RStudio will have the same formatting as the specified style file.
pdf documents
Producing pdf documents requires that LaTeX be installed on your computer
- Few YAML options
- Lots of table options, including kableExtra
- Can use LaTeX code for formatting
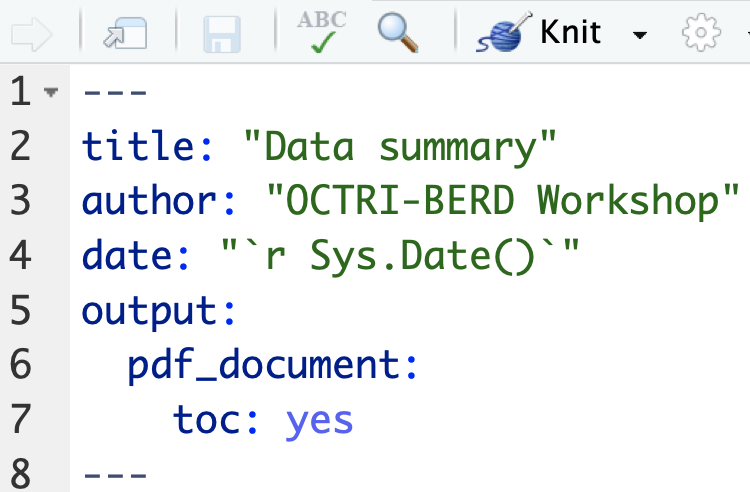
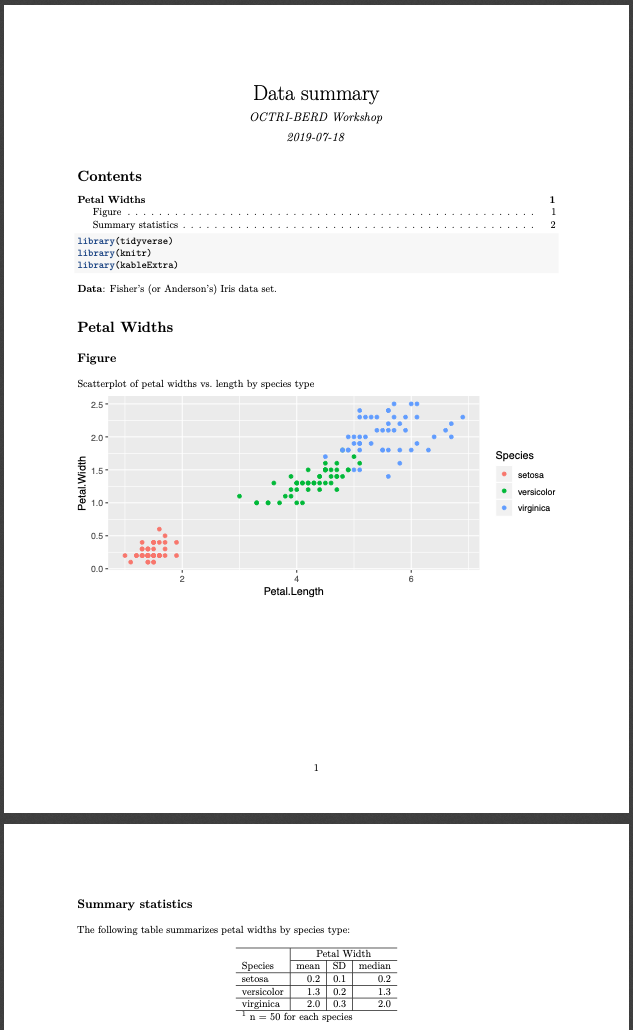
See pdf_example3.Rmd for code and pdf_example3.pdf for output.
Practice!
Change the YAML of example2/example2.Rmd to
- Add your name as author
- Produce a Word document or a pdf document
Extensions and Tips
Real time knitting: xaringan::inf_mr()
Instead of clicking "Knit" every time to see your updated document output, try this:
After installing the xaringan package,
.Rmd files can be run and rendered "live" as you type/save when you either run
xaringan::inf_mr()in the console when your .Rmd file is open. Or, click on on Adddins (top of screen), scroll down to "Xaringan" and click on "Infinite Moon Reader"
This is a new feature, so you need the most recent version of xaringan and RStudio. It works well for html_document output.
Reproducible Workflow
Be Organized
Your files must make sense to yourself 6 months from now, and/or other collaborators.
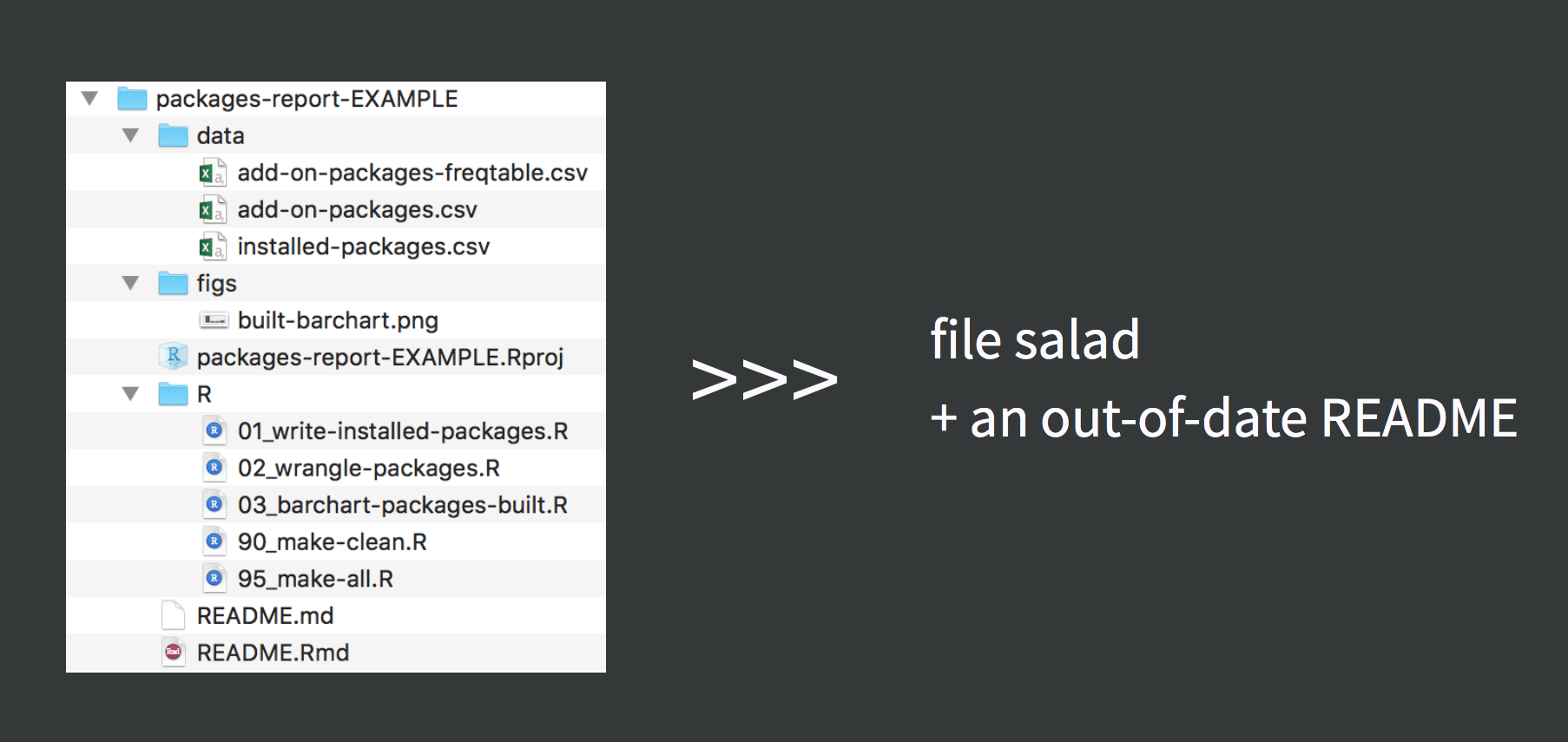
No! Absolute! File! Paths! (don't setwd())
Absolute paths ≠ reproducible
Relative paths = reproducible (if done correctly)

Jenny Bryan's oft quoted opinion; see post on Project-oriented workflow
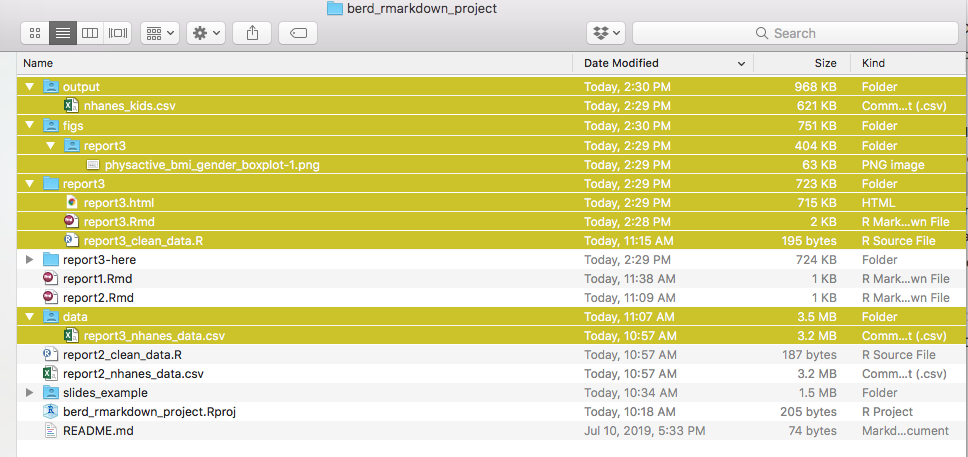
Project directory structure
- The .Rproj file sets your working/home directory (USE PROJECTS)
# Use a relative path, "relative to" the project folderread_csv("mydata.csv") # looks in .Rproj folder- When .Rmd files knit, they look for sourced files in the folder they live in
```{r data, eval=TRUE}read_csv("mydata.csv") # looks in .Rmd's folder```- It's good practice to organize all your code/data/output into separate folders
These three facts together can cause a headache.
- Enter
here::here()!
Everything in one folder
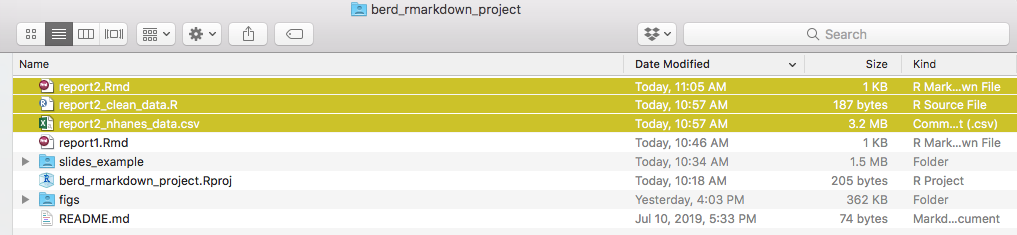
After knitting, this gives you (file 🥗)
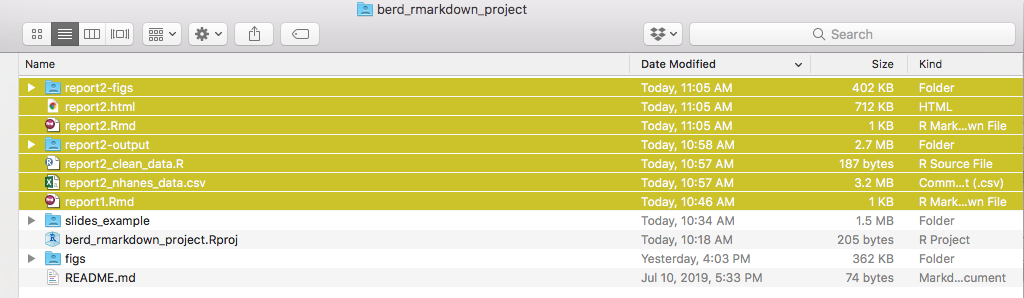
Slightly more organized
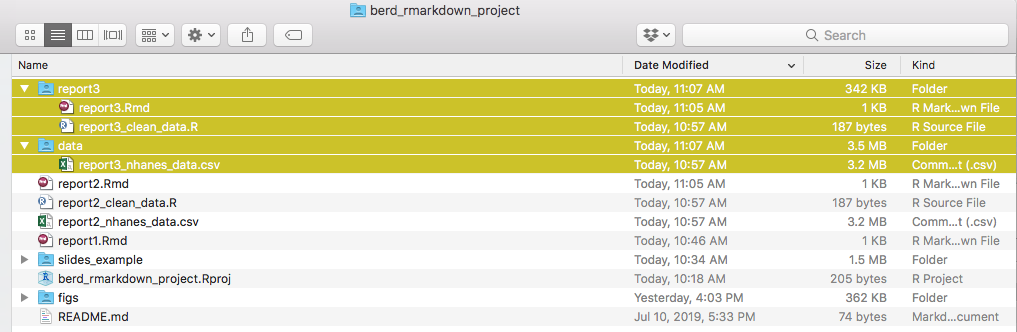
After knitting, this gives you:
Dot dot: A tip about "moving up" a directory/folder
- In unix, to point to the folder one level up (it contains the folder you're in), use
..or../- As in
cd ..moves up one directory, - or
cp ../myfile.txt newfile.txtcopies a file one level up into the current folder (working directory)
- As in
- In
.Rmdwhen you want to source the data in thedata/folder, you could use..to move up a folder into the main directory, and then back down into thedata/folder:
# From the .Rmd folder, move up one folder then down to the data foldermydata <- read_csv("../data/report3_nhanes_data.csv")
here::here() → relative paths to the project directory
- The
herepackage'shere()function solves this issue of inconsistent working directories. - The point of RStudio project workflow is to always have the same "home" working directory = where the
.Rprojfile is. here::here()returns the project directory as a string- Fully reproducible if the whole folder is moved or shared or posted to github
- Portable to ALL systems (Mac, PC, unix), don't worry about
/(Mac) or\(PC) or spaces etc
here::here()[1] "/Users/minnier/Google Drive/BERD R Classes/berd_r_courses_github"here::here() with folders and filenames
here::here("folder","filename")returns the entire file path as a string- These file paths work when running a
.Rmdfile interactively like a notebook, when knitting it, when copying it to the console, wherever, whenever!!
here::here("data","mydatafile.csv")[1] "/Users/minnier/Google Drive/BERD R Classes/berd_r_courses_github/data/mydatafile.csv"here::here("data","raw-data","mydatafile.csv")[1] "/Users/minnier/Google Drive/BERD R Classes/berd_r_courses_github/data/raw-data/mydatafile.csv"We will explore how and when to use this in the exercises.
Practice!
Within your project folder, open this file and follow the instructions:
example4/example4.Rmd
More Extensions and Tips
Even more organized: child documents
If you want to have separate .Rmd files that are sourced in one large document, you can have "child document chunks":
A file called report_prelim.Rmd in the analysis/ folder
(No YAML):
# Details about experimentHere are some details.I can make a plot, too.```{r plotstuff}plot(x,y)```In the main doc main_doc.Rmd
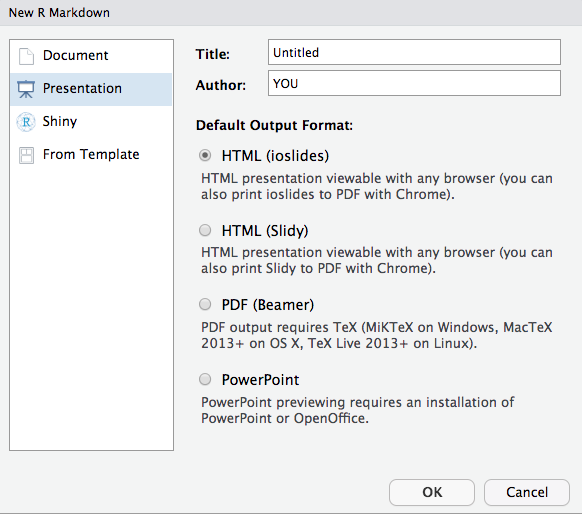
---title: "Main Report:output: html_document---# Preliminary Analysis```{r child = here("analysis","report_prelim.Rmd")}```# Conclusion```{r}kable(summarytable)```Make presentation slides
- These slides were made using a
.Rmdfile with thexaringanpackage! - Simple templates can be found in
File -> new File -> R Markdown -> Presentation
- Each type of presentation uses different syntax to start a new slide, such as
# Slide Header, or---
- ioslides and Slidy are html slides; simple options
- Beamer is from LaTeX
- Xaringan is html based on java script remark.js; has the most flexibility for customizing slides
- PowerPoint is in the newest RStudio release; can use custom templates
Presentations Practice!
Open example4/example4_pres.Rmd and follow instructions.
Bonus: Try using xaringan::inf_mr() to update the output in real time.
Tabsets
A nice feature for showing multiple images or sections is with tabbed sections:
## Results {.tabset}### By Species```{r}ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Species))+ geom_point()```### Panel Species```{r}ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Species))+geom_point()+ facet_wrap(~Species)```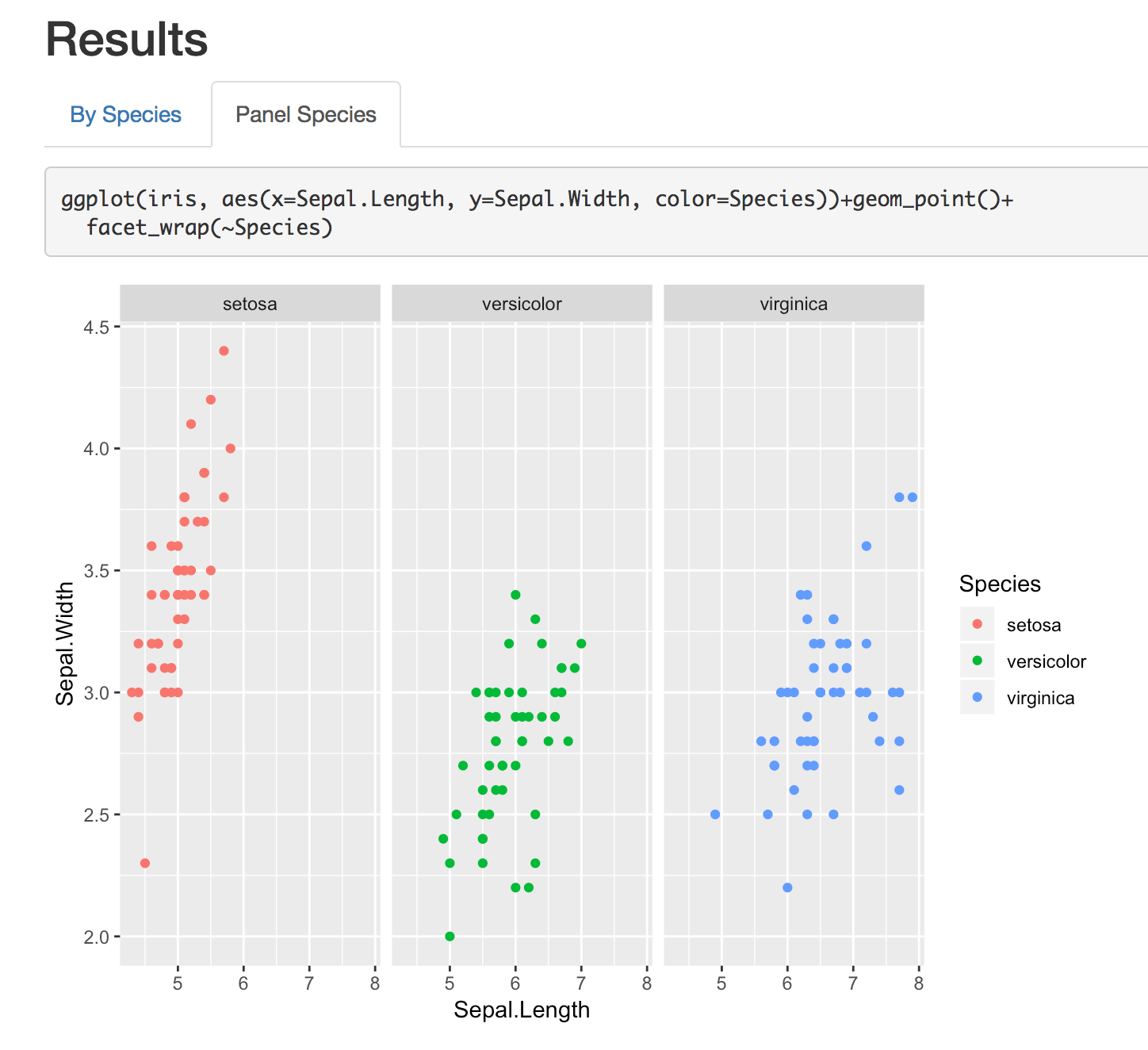
Using other programming languages
- RStudio can run multiple programming languages in the same
.Rmd(if they are installed on the computer), including SAS, STATA, and python. - For more on how to use STATA and SAS, for example, see the documentation for these packages:
names(knitr::knit_engines$get()) [1] "awk" "bash" "coffee" "gawk" "groovy" "haskell" [7] "lein" "mysql" "node" "octave" "perl" "psql" [13] "Rscript" "ruby" "sas" "scala" "sed" "sh" [19] "stata" "zsh" "highlight" "Rcpp" "tikz" "dot" [25] "c" "fortran" "fortran95" "asy" "cat" "asis" [31] "stan" "block" "block2" "js" "css" "sql" [37] "go" "python" "julia" "sass" "scss"Other languages: Limitations
- Each code chunk is run separately as a batch job when using other languages, so it's tricky to pass on objects/data to later code chunks.
- Easy way:
- Use one language to clean data & save the cleaned data as a file
- source the file and continue in another language.
- Other packages can be loaded that help to link objects from various languages, i.e.
- reticulate can store objects created by python code for use in R
- StataMarkdown and SASMarkdown use chunk option
collectcode=TRUEto save code output.
```{r setup} library(SASmarkdown)``````{sas clean_data, collectcode=TRUE}/* clean data with SAS code *//* export to file */``````{sas analyze_data}/* analyze data from above code */``````{r analyze_data}# source clean data file and run code```Knit other types of output
- Journal articles, custom templates
- File → New File → R Markdown → From template
- Dashboards: flexdashboard report output
- Interactive reports with shiny
- Interactive tutorials with learnr
- Websites: blogdown
- Books: bookdown
- Posters: posterdown
- Grad school theses: thesisdown
- It's really endless....
rmarkdown::render()
It can sometimes be easier to set options and change output files/locations when using the render() function in the rmarkdown package. This is also useful for rendering multiple documents in a batch, or using parameterized reports.
In a .R file, or in the console, run commands to knit the documents:
library(rmarkdown)render("report1.Rmd")# Render in a directoryrender(here::here("report3","report3.Rmd"))# Render a single formatrender("report1.Rmd", output_format = "html_document")# Render multiple formatsrender("report1.Rmd", output_format = c("html_document", "pdf_document"))# Render to a different file name or folderrender("report1.Rmd", output_format = "html_document", output_file = "output/report1_2019_07_18.html")knitr::purl() → .R file
Run in the console or keep in a separate R file to extract all the R code into a .R file.
# makes an R file report1.R in same directorknitr::purl("report1.Rmd")# Can be more specific with outputknitr::purl(here::here("report3","report3.Rmd"), # Rmd location out = here::here("report3","report3_code_only.R")) # R output locationknitr::knit_exit(): End document early
- Exit the document early.
- Place this in your
.Rmdto end document there and ignore the rest. - Run parts of the document at a time
```{r}knitr::knit_exit()```Parameterized Reports
---title: My Reportoutput: html_documentparams: data: file.csv printcode: TRUE year: 2018---```{r setup, include=FALSE}knitr::opts_chunk$set( echo = params$printcode)``````{r}mydata <- read_csv(params$data)mydata <- mydata %>% filter(year==params$year)```- Use the Knit button and you will be prompted for values
- Use
rmarkdown::render(default values are set in YAML) - See chapter in R Markdown book for details
rmarkdown::render( "myreport.Rmd", params = list(data = "newfile.csv", year = "2019", printcode = FALSE), output_file = "report2019_newfile.html")Many more bonus tips
- Use git and github for version control, and use output format github_document - see an example
- Quickly convert
.Rfiles to.htmlwith the notebook/compile button orknitr::spin() - Include HTML headers or Latex preambles and files for definitions in YAML
- Add references and a bibliography with BibTex
.bibfiles - Similar to
.Rmdare RStudio "notebooks" -- like an.Rmdbut all the output is saved as it is run in the notebook. - Publish rendered html on Rpubs with Publish button, or through github + netlify.
- Look at these slides by Alison Hill and these by Yihui Xie for many, many more tips and examples
References
- RStudio's R Markdown lessons
- Xie Y. et al R Markdown: The Definitive Guide book online
- Explanation of difference between knitr/Rmd/pandoc
- Teach data science: Getting started with R Markdown
- Alison Hill & Yihui Xie's Advanced R Markdown Workshop Materials
- UCLA's Intro to R Markdown slides
- Software Carpentry Learning R Markdown Materials
Cheatsheets:
Possible Future Workshop Topics?
- tables
- ggplot2 visualization
- advanced tidyverse: functions, purrr (apply/map)
- statistical modeling in R
Contact info:
- Jessica Minnier: minnier@ohsu.edu
- Meike Niederhausen: niederha@ohsu.edu
This workshop info:
- Code for these slides are on github, with links to other course materials: jminnier/berd_r_courses
- The
.Rmdfile that generated the slides is on github and can be downloaded here, though you need to download the whole R project to knit the file. - The project folder of examples can be downloaded at github.com/jminnier/berd_rmarkdown_project & the solutions are in the
solns/folder.